Published online Jul 14, 2007. doi: 10.3748/wjg.v13.i26.3619
Revised: February 18, 2007
Accepted: March 21, 2007
Published online: July 14, 2007
AIM: To analyze the gene expression profiles of mice livers injured by Leigongteng and explore the relationship between the differentially expressed genes and liver damage.
METHODS: The experimental mice were randomly divided into a control group and a liver-injured group in which the mice were administrated 33 μγ of triptolide/kg per day for 30 d. Liver mRNAs were extracted from animals in both groups and were reverse-transcribed to cDNA with dUTP labeled by different fluorescence (Cy3, Cy5) as hybridization probes. The mixed probes were hybridized with oligonucleotide microarray chips. The fluorescent signal results were acquired by scanner and analyzed with software.
RESULTS: Among the 35852 target genes, 29 genes were found to be significantly differentially expressed, with 20 genes up-regulated and 9 genes down-regulated. The reliability of the differentially expressed genes was validated by RT-PCR experiments of 5 randomly selected differentially expressed genes.
CONCLUSION: Based on the biological functions of the differentially expressed genes, it is obvious that the occurrence and development of liver damage induced by Leigongteng in mice are highly associated with immune response, metabolism, apoptosis and the cell skeleton of liver cells. This might be important for elucidating the regulatory network of gene expression associated with liver damage and it may also be important for discovering the pathogenic mechanisms of liver damage induced by Leigongteng.
- Citation: Chen Y, Zhang XM, Han FM, Du P, Xia QS. Gene expression profile analyses of mice livers injured by Leigongteng. World J Gastroenterol 2007; 13(26): 3619-3624
- URL: https://www.wjgnet.com/1007-9327/full/v13/i26/3619.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i26.3619
Tripterygium wilfordii Hook.f., which is called Leigongteng in Chinese traditional medicine, is a type of euonymus plant and is widely used to treat autoimmune diseases, such as rheumatoid arthritis, chronic nephritis, ankylosing spondylitis and various dermatoses. It also has other functions, such as anti-inflammatory, anti-tumor, and anti-fertility functions[1,2], and has special efficacy in ocular diseases, such as White Behcet's disease, Raiter syndrome, scleritis and severe uveitis. Additionally, Leigongteng is promising as an anti-rejection application after organ transplantation.
Leigongteng plays a role in the treatment, to some extent, by inhibiting cell-mediated immune responses.
Triptolide is the main active ingredient of Leigongteng, and also the major toxic component. During the past 50 years, the poisoning incidents caused by Leigongteng and its related drug were reported frequently[3], and the adverse effects were mainly damage to the reproductive system, blood circulation system, digestive system and urinary system. Liver function abnormalities, liver intumescence and an increase in alanine transaminase (ALT) activity were often seen in the poisoned patients. Therefore, Leigongteng's clinical application is limited due to low safety. Liver toxicity associated with Leigongteng has been verified by conventional pharmacological experiments and clinical observations, however, it is still necessary to search for the mechanism of liver injury on a molecular level.
Microarray techniques developed in the last decade with properties of high throughput, minisize and automation, are ideal technical platforms for toxicological research. Microarray technology has been widely used for assessment of environmental harm and drug safety[4-7]. Our group has used microarray techniques to study the gene expression of some typical synthetic chemicals and Chinese traditional drugs, and we have analyzed the correlation between liver injury and differentially expressed genes[8-10]. In the present work, complete mouse genome oligonucleotide microarrays were used to compare the gene expression profiles of mice livers between control and Leigongteng-treated groups. We also analyzed the mechanisms for key differentially expressed genes and liver injury induced by Leigongteng.
Crystal core® CAP-F0006 mice genome oligonucleotide microarrays (35852 Oligo DNA of 70 mer in length, which come from the mice Oligo library of Operon Corporation) and a LuxScan 10KA dual-channel laser scanning device were purchased from CapitalBio Corporation, Beijing. GenePix Pro 4.0 image processing software was purchased from Axon Instruments Corporation. Tripterygium tablets were purchased from 999 Huangshi Pharmaceutical Corporation, China (batch number: 050704, 33 μγ of triptolide in each pill). Assay reagents for determining ALT, aspartate aminotransferase (AST), alkaline phosphatase (AKP), and total protein (TP) were all purchased from Nanjing Jiancheng Bioengineering Institute, China. The Experimental Animal Center of Hubei University permit number for using experimental animals was SYXK 2005-0035.
36 SPF level Kunming mice (male, weight 20 ± 2 g, provided by Hubei Laboratory Research Animal Center, permit number: SCXK 2003-0005) were randomly divided into a Leigongteng-treated group and a control group, each containing 18 mice. The mice of the Leigongteng-treated group were given, by intragastric infusion, the suspended liquid of tripterygium tablet every morning in a dosage of 0.2 mL/10 g body weight (equal to 10 times the adult dose). Each milliliter of the above suspended liquid contained 0.33 μγ triptolide. The mice of the control group were given, by intragastric infusion, the same volume of saline daily. After 30-d administration, 12 mice were randomly selected from each group to withdraw blood samples from the inner palpebrale angle, with the eyeball removed for determining serum ALT, AST and AKP. The liver and spleen indices were calculated, some liver tissues from the left section were taken and fixed in 10% formaldehyde solution for pathological observation, and 1% homogenate of the remaining liver tissues was taken for testing total protein. The other six mice in each group were used for microarray experiments.
RNA extraction and probe preparation: For each of the remaining six mice in each group, 100 mg of liver tissue from the left section was taken to extract total RNA by a modified one-step method[11]. The quality of total RNA was checked by denaturing agarose gel electrophoresis in formaldehyde. The total RNAs were taken equally from each sample and mixed, then reversely transcribed to cDNA, followed by labeling according to the method reported by Schena[12,13]. The labeled cDNAs were used as probes to hybrid with Crystal core® CAP-F0006 mice genome oligonucleotide microarrays. The microarray experiment was an obversely labeled experiment if the probes of the Leigongteng-treating group and control group were labeled with Cy5-dUTP and Cy3-dUTP, respectively.
Otherwise, the microarray experiment was a reversely labeled experiment. Two obversely labeled chips were used for a repeat experiment to avoid false positive results. One reversely labeled chip was used to avoid the deflection of the fluorescence label of Cy5 and Cy3[14].
Microarray hybridization: Labeled probes were isometrically dissolved in 30 μL of hybridization solution (3 × SSC, 0.2% SDS, 5 × Denhardt's solution, 25% formamide), then hybridized with an oligonucleotide microarray chip at 42°C overnight. After hybridization, the oligonucleotide microarray chips were washed first in 0.2% SDS and 2 × SSC at 42°C for 5 m, then in 0.2 × SSC for 5 m at room temperature.
Hybridization signal analysis: GenePix Pro 4.0 software was used to transform the image signals of the chip into digital signals, and then the digital signals were normalized by using the Lowess method[15]. The genes that were compatible with the following three conditions were judged to be differentially expressed genes: (1) signal value ≥ 800, (2) Ratio ≥ 2.0 or ≤ 0.5, and (3) the tendency of gene expression was consistent on the above three chips.
Experimental verification of differentially expressed genes: To verify the authenticity of the differentially expressed genes mentioned above, five of them, such as Sult5a1, Lcn2, Mt1.Clrfa and Hamp, were randomly selected for RT-PCR verification by using GAPDH as a control. The RT-PCR product abundance of each of the mentioned 5 genes was determined by agarose gel electrophoresis.
General Observation: Compared with the control group, the mice of the Leigongteng-treated group were observed to have worse appetites, weaker activities, dark hair, and slower increases in body weight.
Liver index and biochemical assays: Compared with the control group, the liver index, spleen index, and serum ALT, AST, and AKP activities of the mice in the Leigongteng-treated group were significantly increased, and liver TP levels were significantly lower (Table 1).
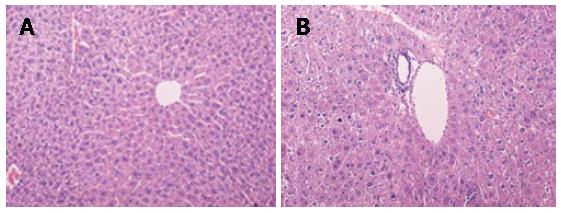
Histopathological observation: Compared with the control group, it was observed in the Leigongteng-treated group that liver cells were extremely swollen, cytoplasts were loosened, cytosolics were colored lightly, the hepatic plates were fractured, hepatic cords were disordered, hepatic cells were fused and dissolved, karyon were condensed, obvious vacuolar and balloon-like changes were observed, and there was also considerable inflammation-related cell infiltration (Figure 1).
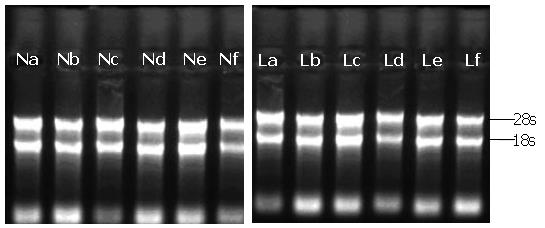
The quality of total RNA: The ratios of A260/A280 in the total RNA ranged from 1.8 to 2.2, indicating the RNA was quite pure. The results of gel electrophoresis are shown in Figure 2. There were two clear bands standing for 28S rRNA and 18S rRNA, which indicated that the extracted mRNA was integral, since the 28S rRNA band was almost two times brighter than the 18S rRNA band.
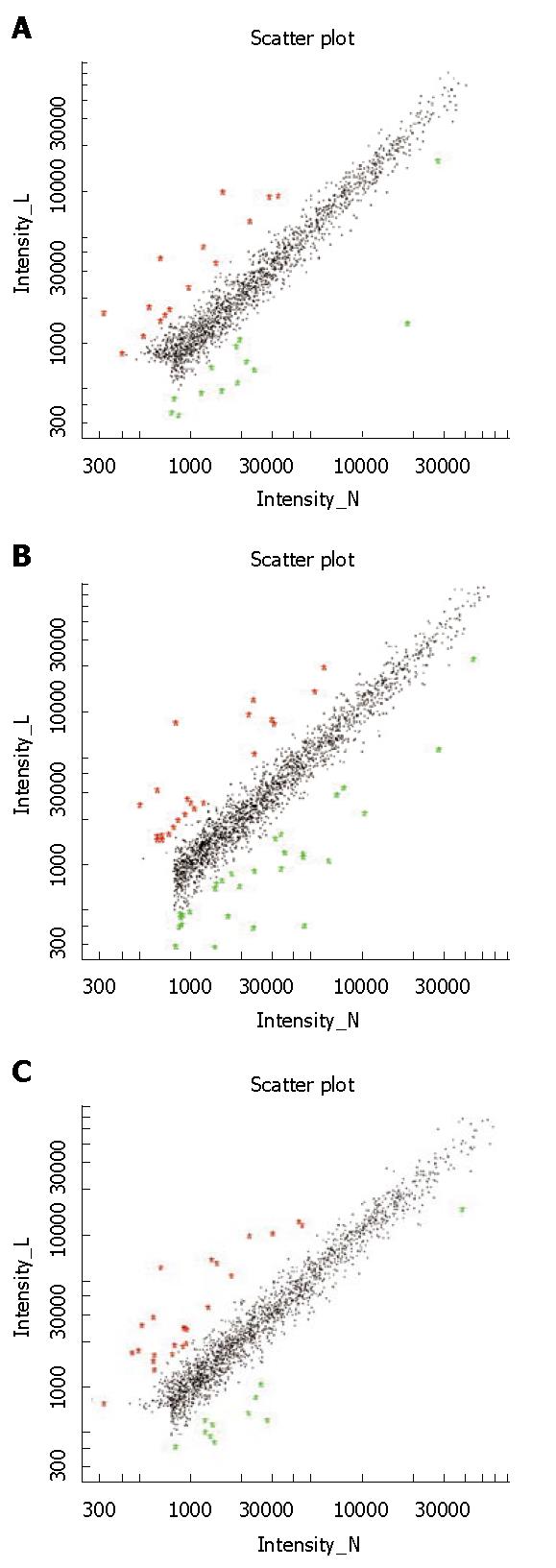
Differentially expressed genes: Figure 3 shows the schematic plots for signals of the chip hybridization experiment. The horizontal and vertical axes represent the fluorescence signals of the control group and the Leigongteng group, respectively. Each spot stands for hybridization signal for each gene in the microarray. The red and green spots distributed in the ranges of slope > 2.0 and of slope < 0.5, representing differentially expressed genes. The black spots distributed in the range of slope 0.5-2, representing genes that were not differentially expressed.
Compared with the control group, there were 29 differentially expressed genes in the Leigongteng-treated group. Among them, 20 genes were up-regulated and 9 genes were down-regulated, and 27 genes were function-known genes, which could be divided into groups according to their biological functions (Table 2).
| Catalog | GenBank access number | Names of the genes | Cy5/Cy3(average) |
| Metabolism | AB059565 | aldo-keto reductase family 1, member C18 (Akr1c18) | 0.226 |
| NM_032541 | hepcidin antimicrobial peptide(Hamp) | 0.354 | |
| L38990 | glucokinase (Gck) | 0.412 | |
| AY273812 | 3-ketoacyl-CoA thiolase B(MGC29978) | 0.445 | |
| NM_013786 | hydroxysteroid (17-beta) dehydrogenase 9 (Hsd17b9) | 2.277 | |
| BC010769 | apolipoprotein A-IV (Apoa4) | 2.759 | |
| BC013476 | cytochrome P450, family 4, subfamily a, polypeptide 10(Cyp4a10) | 2.948 | |
| BC018263 | cis-retinol/3alpha hydroxysterol short-chain dehydrogenase-like(CRAD-L) | 2.988 | |
| CA478631 | metallothionein 1 (Mt1) | 4.404 | |
| BC050123 | asparagine synthetase (Asns) | 5.031 | |
| Immune response | AK005018 | H-2 class II histocompatibility antigen, E-B beta chain precursor | 0.243 |
| BC061489 | Ia-associated invariant chain (Ii) | 0.311 | |
| NM_010738 | lymphocyte antigen 6 complex,locus A (Ly6a) | 2.257 | |
| AA285471 | Lymphocyte antigen Ly-6F.1 precursor | 2.318 | |
| NM_011318 | serum amyloid P-component(Apcs) | 2.657 | |
| M31418 | interferon activated gene 202B(Ifi202b) | 2.823 | |
| NM_009117 | serum amyloid A 1 (Saa1) | 3.763 | |
| BC052495 | Serum amyloid A-1 protein precursor | 5.467 | |
| BG917714 | lipocalin 2 (Lcn2) | 8.587 | |
| NM_011315 | serum amyloid A 3 (Saa3) | 13.1 | |
| Cell skeleton | BI247008 | orosomucoid 2 (Orm2) | 2.132 |
| NM_145594 | fibrinogen-like protein 1 (Fgl1) | 2.823 | |
| Receptors | BC054522 | transferrin receptor (Tfrc) | 2.817 |
| Apoptosis | AK051011 | DNA fragmentation factor alpha subunit (DNA fragmentation factor 45 kDa subunit) (DFF-45) | 4.042 |
| U44088 | pleckstrin homology-like domain, family A, member 1 (Phlda1) | 0.297 | |
| NM_011817 | growth arrest and DNA-damage-inducible 45 gamma (Gadd45g) | 0.214 | |
| Cancer related genes | BC050858 | RasGEF domain family, member1B (Rasgef1b) | 2.311 |
| Unknow | NM_183257 | RIKEN cDNA 1810073K19 gene (1810073K19Rik) | 0.368 |
| NM_025326 | RIKEN cDNA 0610011I04 gene (0610011I04Rik) | 2.421 |
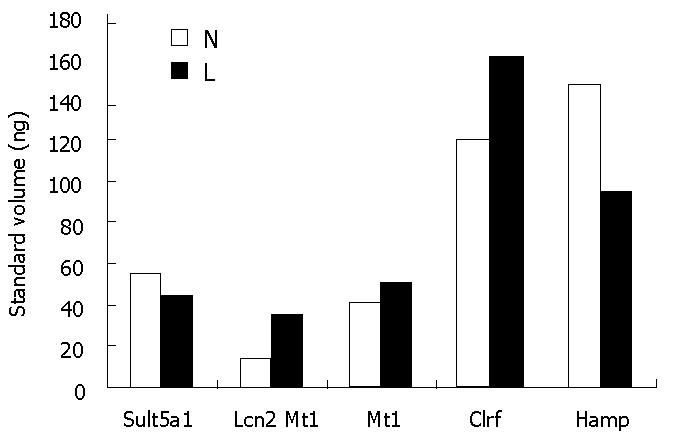
According to the results of RT-PCR experiments, the abundances of five randomly selected differentially expressed genes (Sult5a1, Lcn2, Mt1, Clrf, Hamp) are shown in Figure 4. Based on the results of the chip experiment, Lcn2, Mt1, and Clrf were significantly up-regulated, while Sult5a1 and Hamp were significantly down-regulated in the Leigongteng-treated group. The RT-PCR experiment showed that the change tendencies were consistent in the 5 genes. These results verified that the genes identified in the chip experiments were differentially expressed.
Assessing the pre-clinical safety of Chinese traditional medicine is an important topic that needs to be resolved in modern Chinese medicine. The liver is the most important organ for the metabolism of drugs and xenobiote, and also is an important target organ affected by toxicants. Hence, the toxicity of Chinese traditional medicine on the liver has significant meaning in pre-clinical safety assessments. The efficacy and toxic side effects of Chinese traditional medicines are inevitably related to key gene expression or gene network regulation either directly or indirectly. Exceptional changes in mRNA levels of the related functional genes in specific cells should be very valuable in assessing cell toxicity and toxic mechanisms associated with Chinese traditional medicine.
This study indicated that Leigongteng has an important impact on functional gene expression in mice liver. Here we focused on the correlation between differentially expressed genes and liver injury associated with Leigongteng.
Compared to the control group, there were 10 differentially expressed genes related to metabolism in the Leigongteng-treated group. Among them, genes AB059565, L38990, AY273812 and NM_032541 were down-regulated, while genes NM_013768, BC010769, BC013476, BC018263, CA478631 and BC050123 were up-regulated. According to the literature, exogenous drugs or toxic injury can lead to lipid peroxidation (LPO) of liver cells and damage to mitochondrial function, ultimately leading to cell swelling, degeneration and necrosis. Metallothionein protein is mainly synthesized in the liver and has very high clearance ability on O2 radicals and H radicals, and stress reaction is the key factor in controlling the expression of metallothionein protein[16]. The up-regulated expression of the metallothionein protein gene (Mt1), labeled CA478631, indicated that there was a resistance response to oxidative damage in injured liver cells. Glucokinase is a type of speed limit enzyme regulating glucose metabolism to maintain the steady-state of blood glucose. The glucokinase gene (Gck), labeled L38990, was down-regulated. 3-ketoacyl-CoA thiolase B is a type of mitochondrial enzyme that regulates the metabolism of fatty acid and the oxidation of glucose. The gene of 3-ketoacyl-CoA thiolase B (MGC29978), labeled AY273812, was also down-regulated. The down-regulated expression of these genes that are related to glucose metabolism may be one of the reasons for mice growth slowness induced by administrating Leigongteng. Apolipoprotein A-IV, encoded by gene Apoa4 labeled BC010769, is the ligand of a high-density lipoprotein receptor and it plays an important role in maintaining the structure of lipoprotein and lipid transfer in the process of lipid metabolism. The up-regulated expression of BC010769 may be responsible for stress and regulatory feedback in the injured liver. Aldo-keto reductase family 1, number C18, encoded by gene AB059565, is an important enzyme in lipid metabolism. The down-regulated expression of AB059565 indicates that there was metabolic inhibition of fatty acid in the injured liver, and this may be closely related to the fatty degeneration shown in the liver biopsy. The up-regulated expression of hydroxysteroid (17-beta) dehydrogenase 9 gene (Hsd17b9), labeled NM_013786, is commonly found in liver cancer and is caused by a variety of toxicants[17]. Its up-regulated expression in the Leigongteng-treated group indicates that there was risk of carcinogenesis in the liver. Hepcidin antimicrobial peptide is an extremely important iron-regulating hormone that controls intestinal iron absorption and iron homeostasis in vivo. The down-regulated expression of hepcidin antimicrobial peptide gene (Hamp), labeled NM_032541, suggested that the liver injury may be associated with a metabolic disorder of iron. In addition, the cytochrome P450 gene (BC013476), short-chain dehydrogenase gene (BC018263), metallothionein gene (CA478631) and asparagine synthetase gene (BC050123) were all up-regulated. The abnormal expression of the above 10 genes indicates that metabolism in liver cells was disturbed in the Leigongteng-treated group.
There are also 10 differentially expressed genes associated with immune processes. Among them, the genes of lymphocyte antigen 6 complex, locus A (NM_010738), lymphocyte antigen Ly-6F.1 precursor (AA285471), interferon activated gene 202B (M31418), like protein amyloid P component (NM_011318, NM_009117, NM_011315, BC052495), lipocalin 2 (BG917714) were all up-regulated, while the genes of Ia-associated invariant chain (Ii), labeled BC061489, and H-2 class II histocompatibility antigen, labeled AK005018, were down-regulated. The up-regulated expression of the above 8 genes is associated with inflammatory reactions in liver tissue, which is also the detoxic emergency response induced by the administration of Leigongteng. Therefore, the inflammation of liver cells may be an important cause of the damage induced by Leigongteng in mice liver[18]. The down-regulated expression of Ii and H-2 genes was related to the injury effect of immune mechanisms caused by drugs. The major histocompatibility complex of mice (MHC), encoded by the H-2 gene and expressed by antigen presenting cells[19], can activate the immune response by presenting antigen to T helper cells and this plays an important role in the identification and removal of the external and internal antigen[20]. The down-regulated expression of MHC classI antigen genes is a common mechanism that allows tumor cells to escape from the destruction of anti-tumor T cells. Therefore, the down-regulated expression of H-2 genes indicated that there were T cell-mediated immunological abnormalities in the Leigongteng-treated group.
Both cytoskeleton-associated genes, BI247008 and NM_145594, were up-regulated in the Leigongteng-treated group, and were considered to be an important cause of liver cell degeneration, osteoporosis, cytoplasm and cytoskeletal structure damage. The transferrin receptor, as a membrane protein associated with iron transfer, is widely distributed on the cell membrane. The up-regulated expression of Tfrc, labeled BC054522, may be responsible for the external toxic invasion of liver cells caused by the administration of Leigongteng. The Ras oncogene is considered to be a normal housekeeping gene controlling cell proliferation and differentiation. The activation of this gene can lead to abnormal expression of the encoded products and can trigger malignant transformation of normal cells, which ultimately leads to the occurrence of malignant tumors. The RasGEF domain family member 1B (Rasgef1b), labeled BC050858, is a cancer gene, and excessive expression of BC050858 indicated that there was a high risk of liver cancer in the Leigongteng-treated group. DNA fragmentation factor (DEF), as a main apoptotic signaling pathway, plays an important role in apoptosis (Liu, 2003). The markedly excessive expression of DFF45 (AK051011) in the Leigongteng-treated group showed that DEF45 created an active defense reaction by removing injured cells to maintain the tissue stability. In the formation of liver tumors, the lack of apoptosis of malignant cells is considered to be a decisive factor in tumor development. The deficient expression of apoptosis related genes, such as the nucleoprotein gene Phlda1(U44088) and the growth arrest and DNA-damage-inducible 45 gamma gene (NM_011817), indicate that there was potential risk of cancer for the mice treated with Leigongteng.
In addition to the above function-known genes, there were two function-unknown differentially expressed genes in the Leigongteng-treated group. The relationship between these genes and liver injury is unknown.
| 1. | Chang WT, Kang JJ, Lee KY, Wei K, Anderson E, Gotmare S, Ross JA, Rosen GD. Triptolide and chemotherapy cooperate in tumor cell apoptosis. A role for the p53 pathway. J Biol Chem. 2001;276:2221-2227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 134] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 2. | Huynh PN, Hikim AP, Wang C, Stefonovic K, Lue YH, Leung A, Atienza V, Baravarian S, Reutrakul V, Swerdloff RS. Long-term effects of triptolide on spermatogenesis, epididymal sperm function, and fertility in male rats. J Androl. 2000;21:689-699. [PubMed] |
| 3. | Mei Z, Li X, Wu Q, Hu S, Yang X. The research on the anti-inflammatory activity and hepatotoxicity of triptolide-loaded solid lipid nanoparticle. Pharmacol Res. 2005;51:345-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 101] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 4. | Pennie WD, Tugwood JD, Oliver GJ, Kimber I. The principles and practice of toxigenomics: applications and opportunities. Toxicol Sci. 2000;54:277-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 57] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 5. | Newton RK, Aardema M, Aubrecht J. The utility of DNA microarrays for characterizing genotoxicity. Environ Health Perspect. 2004;112:420-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 71] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 6. | Lutz W, Kur B. Toxicogenomics. New perspectives for the molecular toxicology. Med Pr. 2004;55:193-202. [PubMed] |
| 7. | Hamadeh HK, Bushel PR, Jayadev S, DiSorbo O, Bennett L, Li L, Tennant R, Stoll R, Barrett JC, Paules RS. Prediction of compound signature using high density gene expression profiling. Toxicol Sci. 2002;67:232-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 151] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 8. | Chen Y, Cheng M, Xia QS, Du P. Changes of gene expression profiles in CCl4 injured liver of mice. YaoXue XueBao. 2005;40:898-902. |
| 9. | Han F, Cheng M, Xia Q, Chen Y. Gene expression profile in immunologically injured liver cell of mice. Sci China C Life Sci. 2006;49:454-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 10. | Chen Y, Cheng M, Han FM, Du P. Comparing studies on the differences of CCl4 liver injury and immunity liver injury mice models. Shiyan Shengwu Xuebao. 2005;38:417-422. |
| 11. | Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40518] [Cited by in RCA: 39200] [Article Influence: 1005.1] [Reference Citation Analysis (0)] |
| 12. | Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA. 1996;93:10614-10619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1087] [Cited by in RCA: 959] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 13. | Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6477] [Cited by in RCA: 5130] [Article Influence: 165.5] [Reference Citation Analysis (0)] |
| 14. | Weigelt B, Glas AM, Wessels LF, Witteveen AT, Peterse JL, van't Veer LJ. Gene expression profiles of primary breast tumors maintained in distant metastases. Proc Natl Acad Sci USA. 2003;100:15901-15905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 316] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 15. | Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2380] [Cited by in RCA: 2439] [Article Influence: 101.6] [Reference Citation Analysis (12)] |
| 16. | Mehta A, Flora SJ. Possible role of metal redistribution, hepatotoxicity and oxidative stress in chelating agents induced hepatic and renal metallothionein in rats. Food Chem Toxicol. 2001;39:1029-1038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 17. | Zeindl-Eberhart E, Klugbauer S, Dimitrijevic N, Jungblut PR, Lamer S, Rabes HM. Proteome analysis of rat hepatomas: carcinogen-dependent tumor-associated protein variants. Electrophoresis. 2001;22:3009-3018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 18. | Decker K. Biologically active products of stimulated liver macrophages (Kupffer cells). Eur J Biochem. 1990;192:245-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 660] [Cited by in RCA: 629] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 19. | Garrido F, Cabrera T, Concha A, Glew S, Ruiz-Cabello F, Stern PL. Natural history of HLA expression during tumour development. Immunol Today. 1993;14:491-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 347] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 20. | Marincola FM, Jaffee EM, Hicklin DJ, Ferrone S. Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv Immunol. 2000;74:181-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 839] [Cited by in RCA: 825] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
S- Editor Liu Y L- Editor Lutze M E-Editor Wang HF