Published online Mar 28, 2007. doi: 10.3748/wjg.v13.i12.1799
Revised: November 17, 2006
Accepted: February 8, 2007
Published online: March 28, 2007
AIM: To clone human liver special F protein and to express it in a prokaryotic system.
METHODS: Total RNA was isolated from human liver tissue and first-strand cDNA was reverse transcribed using the PCR reverse primer. Following this, cDNA of the F protein was ligated into the clone vector pUCm-T. The segment of F protein’s cDNA was subcloned into the expression vector pET-15b and transformed into E. coli BL21 (DE3) pLyss. Isopropy-β-D-thiogalactoside (IPTG) was then used to induce expression of the target protein.
RESULTS: The cDNA clone of human liver special F protein (1134bp) was successfully produced, with the cDNA sequence being published in Gene-bank: DQ188836. We confirmed the expression of F protein by Western blot with a molecular weight of 43 kDa. The expressed protein accounted for 40% of the total protein extracted.
CONCLUSION: F protein expresses cDNA clone in a prokaryotic system, which offers a relatively simple way of producing sufficient quantities of F protein and contributes to understanding the principal biological functions of this protein.
- Citation: Liu SY, Yu XD, Song CJ, Lu W, Zhang JD, Shi XR, Duan Y, Zhang J. Cloning and expression of special F protein from human liver. World J Gastroenterol 2007; 13(12): 1799-1804
- URL: https://www.wjgnet.com/1007-9327/full/v13/i12/1799.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i12.1799
F protein was first isolated from mouse liver by Fravi and Lindenmann in 1968[1]. This particular protein can be divided into F1 and F2[2]. Liver disease is the most serious disease impairing public health. Although there are many diagnostic methods to estimate the severity of injury to the liver, none has been demonstrated to be completely reliable. It is therefore necessary to develop serological test methods that can precisely predict the degree of damage to liver. F protein is mainly concentrated in hepatic cytoplasm where its expression is 1370-fold higher than in serum. However, when liver cells are damaged, F protein is released to increase its serum concentration. It is therefore possible to establish a correlation between the concentration of serum F protein and the degree of hepatocellular damage.
Using affinity-based purification techniques, Oliveira and colleagues[3,4] were the first to describe the existence of F protein in mammalian cells. F protein is a substance within the liver that identifies organ specificity and genus non-specificity. A large amount of F allelic protein exists in the liver of rats. Even though its biological function is unclear, it is thought to be responsible for immune tolerance and immune response observed in rats following transplantation[5-11].
At present, only a few laboratories have begun to elucidate the proper biological function of F protein[5,6,10]. Indeed, no database contains the cDNA sequence of human variant of F protein. Here, we describe the cloning and expression of human F-protein from cDNA prepared from human liver[12].
Clone receptor E. coli DH5a was purchased from Dalian BAO and kept in our laboratory. Clone vector pUCm-T was purchased from Shanghai Shenggong Company. The expression vector pET-15b was provided by Professor Lu Yichen. AMV RNA reverse transcriptase, endonuclease BamHI, EcoRI, NdeI, Taq DNA polymerase (EX -Taq premix), X-gal, isopropy-β-D-thiogalactoside (Pharmacia), DL2000 DNA marker, DL15 000 DNA maker, λDNA/EcoRI + Hind III were all purchased from Dalian BAO. The primers used for PCR were synthesized by Beijing AOKE. Ampicillin was purchased from Huamei and chloramphenicol was purchased from Beijing DINGGUO.
Isolation and detection of total RNA from human liver: RNA was extracted from approximately 0.4 g of fresh liver tissue using 4 mL Trizol® reagent. The purity and integrity of RNA obtained were determined by agar gel electrophoresis.
RT-PCR amplification of human liver F protein cDNA: Using the cDNA sequence of rat F protein from Gene-Bank, primers were designed to amplify the cDNA of F protein by RT-PCR. The forward primer sequence is 5’ P-TCC AGA CCT CAC ACCGTT-3’ and the reverse primer sequence is 5’-TCC AGA CCT CAC ACC GTT-3’. Prior to amplifying the template RNA, cDNA was produced by reverse transcription at 42°C for 60 min, and at 94°C for 5 min. The products were then amplified for 35 cycles at 94°C for 5 min, at 94°C for 30 s, at 55°C for 30 s, at 72°C for 1.5 min, and a final extension at 72°C for 10 min. The PCR products were then analyzed by agarose gel electrophoresis[13].
Insertion of F gene clone into T-carrier: The PCR products of F protein were dissolved in 6 μL of sterilized water under an ultra-violet lamp. DNA fragments were incorporated into the pUCm-T vector using a T4 DNA ligase at 16°C overnight.
Transformation and identification of recombinant bacteria: Calcium chloride treatment rendered DH5a bacteria competent for uptake of the pUCm-T vector. Colonies positive for the exogenous gene were screened using the blue–white patch method, then isolated and named pUCm-T-F. Isolated colonies were digested with NdeI and BamHI and sequenced in Shanghai Shenggong Company with ABI PRISM 377-96.
Construction of expression vector: Plasmid pUCm-T-F was digested using NdeI and BamHI and ligated with the vector overnight at 16°C. Following this the recombinant plasmid was transformed into E. coli, BL21 (DE3) pLyss. The transformed bacteria were selected base on ampicillin and chloramphenicol resistance. Recombinant plasmids were analyzed to confirm possession of vector and termed pET15b-F.
Expression of F protein in bacterial strain: BL21 (DE3) pLyss containing plasmid pET15b-F was grown overnight in LB media at 37°C until the OD was between 0.4 and 0.6. At this time point, IPTG (1 mmol/L) was added to the media and maintained at 37°C for 3 h. Culture media were isolated and expressed proteins were analyzed by Western blot. The blank strain BL21 (DE3) pLyss containing no pET15b-F expression plasmid and the strain containing the blank vector pET-15b (i.e no F-protein) were used as controls.
Purity of expressed protein: Following precipitation and centrifugation the protein was purified by using a Ni-charged IDA agarose affinity chromatography column.
Detection of expressed protein SDS-PAGE: Electr-ophoresis and Western-blot were used to analyze the expression of all proteins obtained in this study.
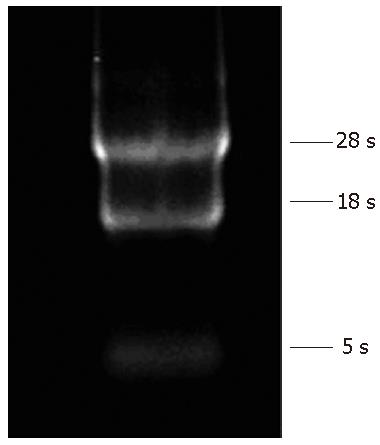
Gel-PAGE of total RNA from human liver tissue: Following isolation of RNA, its integrity and purity were confirmed by agar gel electrophoresis.
As can be seen in Figure 1, lines corresponding to 28s and 18s rRNA bands gave a ratio of 2:1 for 28S and 18S, indicating that no degradation of RNA occurred.
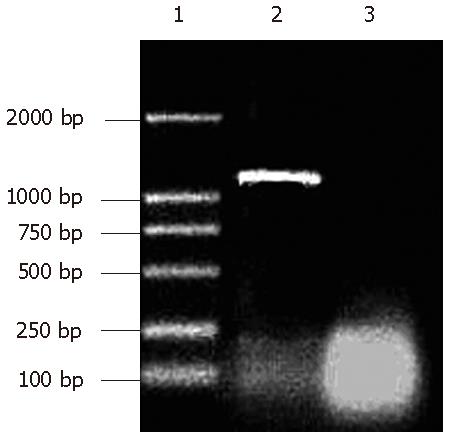
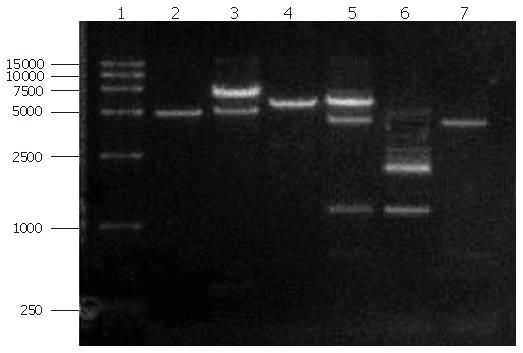
RT-PCR: The isolated RNA was then subjected to RT-PCR with amplified DNA measured by agar gel electrophoresis. The marker used is DL-2000, each strip correlated 2000 bp, 1000 bp, 750 bp, 500 bp, 250 bp, 100 bp in the electrophoresis diagram.
As can be seen from Figure 2, the PCR product was approximately 1.2 kbp, similar to the size of rat F protein as published in Gene Bank.
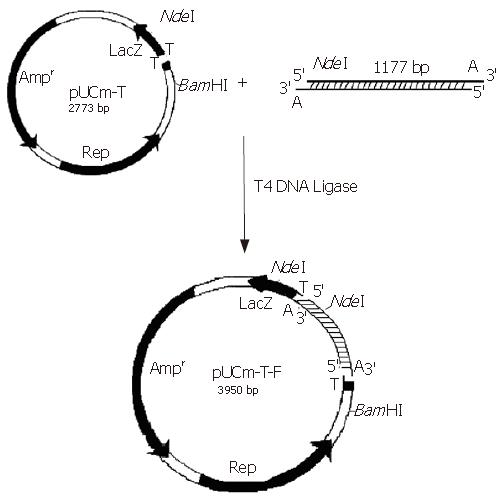
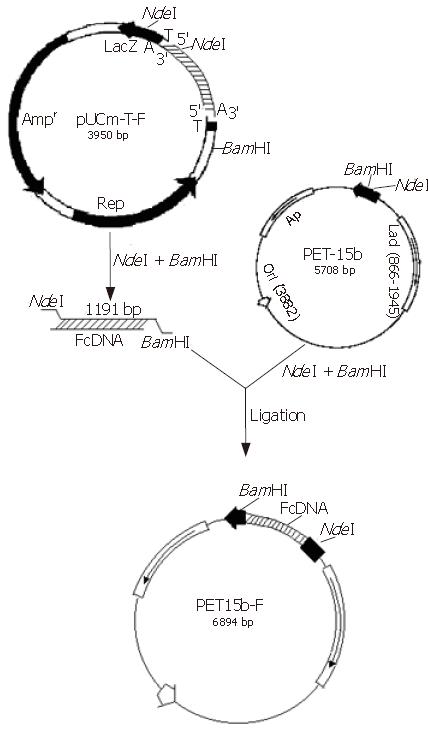
The PCR products were separated on agar gel, extracted, purified and ligated with the pUCm-T vector (Figure 3). The plasmid vector was then digested using NdeΙ and BamHΙ and detected on agarose gel in order to analyze fragment sizes (Figure 4).
The empty vector of pUCm-T was 2773 bp with PCR product being 1177 bp. Therefore, when digested with BamHI, there should be a fragment of 3950 bp. From the polyclonal site of the pUCm-T vector, the BamHI enzyme digestion site was 14 bp downstream of the PCR product insertion place. The EcoR1 site was 84 bp downstream of the PCR product insertion point. Therefore, as can be seen from Figure 4, when both BamHI and EcoR1 were used, fragments of 2675 bp and 1275 bp could be obtained. Sequencing results: The sequencing results were against that of rat F-protein in Genebank with homology to the sequence of human F-protein, confirming the obtained cDNA clone of human liver F protein.
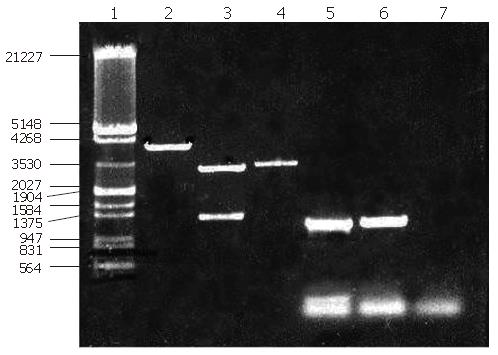
Construction and identification of expression vector: The expressing vector was digested using NdeΙ and BamHΙ, then ligated with the cDNA fragment of F protein (1.3kb). After analysis through digestion, the expression vector was termed pET15b-F (Figure 5).
Digestion and identification of expression vector: The expression strain BL21 (DE3) pLyss contains two plasmids, one is the expression vector pET15b-F and the other is a helper plasmid. Following isolation of these plamids from the expression strain, they were digested with HindIII. Running the digestion products in an agarose gel revealed two bands (Figure 6), one (6891 bp) corresponding to the expression vector containing the exogenous gene encoding for F-protein, the other (5000 bp) to the helper plasmid pLyss. Double digestion (using BamHI and NdeI) of the helper plasmid again yielded a single band as expected whereas double digestion of the expression vector yielded 2 bands, one corresponding to the cDNA of F protein and the other to the cloning vector.
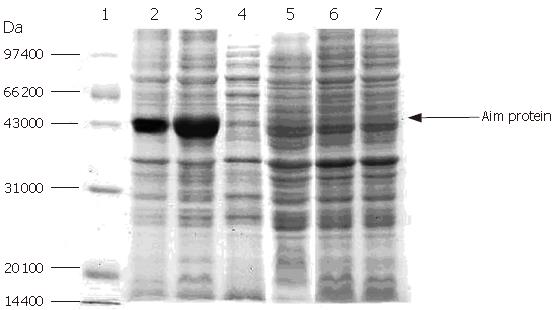
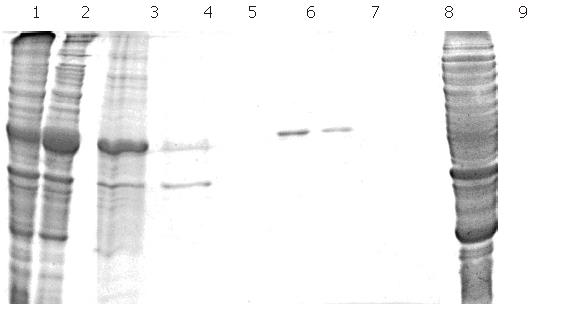
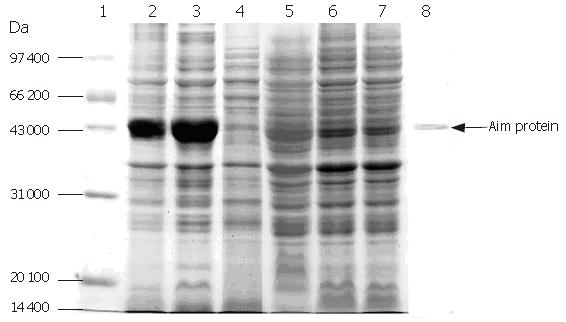
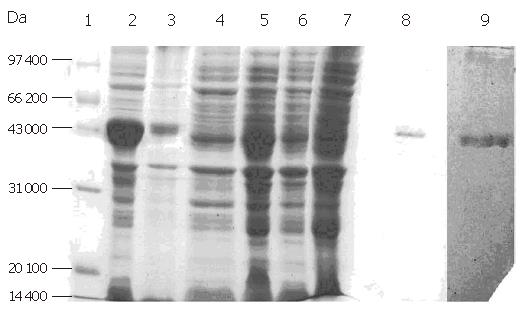
Expression of F protein: After expression of F-protein was induced using 1 mmol/L IPTG, the media were subjected to centrifugation in order to collect all secreted proteins, which were then analyzed using SDS-PAGE (Figure 7). The collected proteins were then purified using a Ni-charged IDA affinity chromatography column and the purified proteins were detected by SDS-PAGE (Figure 8).
As can be seen from Figure 8, there was a substantial amount of rough protein in the preparation prior to purification by chromatography. Following this purification step, pure F protein was obtained.
Electrophoresis of target protein in all expressed and purified strains: The purified target proteins, expression strain, blank control and protein marker were put into SDS-PAGE to detect the size of target protein by electrophoresis. Figure 9 shows 43 kDa of target F protein band.
Western blot analysis of F-protein: Western blot showed strong immunoreactivity by using a polyclonal antibody against human liver F-protein (Figure 10).
Since liver specific F protein was isolated in 1960s[1], it has been used in the diagnosis of hepatic diseases and in judging the degree of hepatocellular damage. Serum F protein has been shown to have a close correlation with the degree of hepatocellular damage[2-4], thus representing a new sensitive and specific serologic marker which could reflect the degree of hepatic damage. No age or sex-related differences have been found in serum F protein levels. Use of serum F protein as a tool to identify liver damage is of great importance[1-4,14].
The importance to study F-protein function is highlighted by reports describing it as an organ specific antigen[4]. It has been previously demonstrated that liver F protein is directly involved in the immunorejection of liver transplants in rats[10,15]. Thymus inoculation of liver F protein has been shown to alleviate the rejection of liver transplants, demonstrating that F-protein is an important antigen against transplantation[10,15]. Due to the difficulties in separating and purifying tissue cells, the study of tissue specific antigenicity in this regard has been hindered. As studies on the expression and distribution of liver F protein are limited, it is difficult to perform a detailed analysis of its function. The Mrs of F protein have been reported to be 40 kDa-45 kDa[16], which is possibly due to the differences in techniques used to isolate F protein[16]. Due to the difficulties in obtaining sufficient amount of F protein, it is difficult to show its proper biological function.
At present there are no published studies on the clone of human liver F-protein gene. No corresponding gene sequence has been reported. Due to the similarity between the genes of rodents and humans and the fact that no published sequence of human liver F-protein is available, we used the published rodent gene sequence to design 10 pairs of forward primers and reverse primers. We subsequently applied them to the RNA isolated from human liver tissue. The cDNA obtained from the subsequent RT-PCR was then subjected to PCR amplification with products analyzed by agarose gel electrophoresis. The 10 pairs of forward primers and reverse primers are as follows:
1 F primer: 5’-CGG CAT ATG TAC TGG GAC AAA GGA CCA AAG CCT GAG AGA-3’
R primer: 5’-TCC AGA CCT CAC ACC GTT GGT CTC CAG-3’
2 F primer: 5’-CTG GAT CCG GAG GTT TGA CTA AGA TCA ATC A-3’
R primer: 5’-GCG AAT TCT GGA TCT GGG ACT TCT TCT TG-3’
3 F primer: 5’-CTG GAT CCG GTG CAC ACG GAA TAT AGC TC-3’
R primer: 5’-GCG AAT TCT TTA ATC GGG AGG GCT GGA-3’
4 F primer: 5’-TAT CAT ATG GTA CTG GGA CAA AGG ACC AAA GCC T3’
R primer: 5’-TAT GGA TCC CAT CTG GGA AGG CAT CTT TAC TCC A3’
5 F primer: 5’- CAG GCT GCC TCC TTC TAT TG-3’
R primer: 5’- CAG GTG GTC ACC CAT CTC TT-3’
6 F primer: 5’-AAG AGA TGG GTG ACC ACC TG-3’
R primer: 5’-AGC ACA GCG AAC TTC ACC TT-3’
7 F primer: 5’-GTA CTG GGA CAA AGG ACC AAA GCC-3’
R primer: 5’- GAT CTG GGA AGG CAT CTT TAC TCG –3’
8 F primer: 5’-TCC CTG GAA CAA GAG ATG G-3’
R primer: 5’-AGC ACA GCG AAC TTC ACC T-3’
9 F primer: 5’-AAG AGA TGG GTG ACC ACC TG-3’
R primer: 5’-TTC ACC TTC CCG AAT TTG TC-3’
10 F primer: 5’-TCC CTG GAA CAA AGA GAT GG3’
R primer: 5’-TTC ACC TTC CCG AAT TTG TC3’
Using these primers we found that only certain primer sequences could result in amplification. Therefore, we selected the pair of forward and reverse primers to inverse transcript and amplify human liver F-protein cDNA resulting in RT-PCR products of 1200 bp.
As the size of the cDNA is so large, after ligating the DNA with the cloning vector a certain number of base mutations occurred. In order to correct them, the following primers were designed to repair these mutations:
1 F primer: 5’-CATATCCTACTGGGACAAAG -3’
R primer: 5’-CTTGATGACATGACTGACCACCTC-3’
2 F primer: 5’-TGGTCAGTCATGTCATCAAGCAAGGG-3’
R primer: 5’-TTCCCTCAAGTGGCGGATCGTTGTG-3’
3 F primer: 5’-CGATCCGCCACTTGAGGGAACGAC-3’
R primer: 5’-GGATCCGATCTGGGAAGGCATC-3’
Finally, it was possible to isolate the PCR products with these three pairs of primers and we used the 1 up stream and the 3 down stream as the primers to amplify the whole sequence. After ligating the PCR products with the cloning vector, the sequencing results from PCR confirmed isolation of the target gene.
In order to optimize protein expression, we carried out experiments under certain conditions that may influence the expression of F protein. The optimal exogenous protein expression occurred with an E. coli OD600 value of 0.3-0.5 when E. coli was in a state of logarithmic growth. In this particular stage the expression of proteins could be influenced by exogenous factors that may promote protein expression. If the OD600 value exceeds 0.6, the growth of E. coli enters the telophase stage whereby a number of factors can negatively influence protein expression, including accumulation of toxic products such as organic aids, H2O2 and decreased pH. Essentially, bacterial reproduction decreases while their mortality increases, so the ability to express exogenous protein significantly decreases. In the exponential growth phase, protein expression is induced by IPTG at the concentration of 0.8-1.2 mmol/L. If the concentration range of IPTG exceeds 0.8-1.2 mmol/L, it promotes maximum protein expression in a period of 5 h. In summary, the optimal experimental conditions for inducing maximum expression of proteins were as follows: E. coli optimum concentration is OD600 = 0.5, Optimum concentration of inducer IPTG is 1.0 mmol/L, and Optimum time is 5 h.
Following submission of the gene sequence of human liver F protein cDNA to GENEBANK (the submission number was DQ188 836), we proceeded to blast the sequence and discovered a 99% sequence homology between human and rat liver F-protein. The bases at 179, 830 and 1133 of human liver F-protein gene sequence was G, T, A and the corresponding base sequence of rat liver F-protein gene sequence was A, C, T, respectively.
Western-blot analysis of the obtained protein revealed that its molecular weight is in agreement with the published molecular weight of human liver F-protein (approximately 43 kDa). Therefore, we could express F-protein and confirm its identity in a prokaryotic system, thus laying a foundation for subsequent studies on elucidating the proper biological function of F-protein.
In conclusion, we report the isolation of total RNA from the rat liver obtaining the intact sequence of F protein cDNA through reverse transcription. The PCR products can be cloned into pUCm-T, confirming that the cloned fragment is identical to that of the rat liver F-protein mRNA sequence. F protein is expressed in an E.coli expression system. Furthermore, the protein has the same immune specialty as human liver F protein.
The authors thank Professor Lu Yi-Chen for providing expression vector pET-15b and Professor Cai Lao-Shi (Academy of Medical Sciences, Blood Institute) for F protein purification.
| 1. | Fravi G, Lindenmann J. Induction by allogeneic extracts of liver-specific precipitating autoantibodies in the mouse. Nature. 1968;218:141-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 41] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 2. | Mori Y, Iwama S, Mori T, Ueda S, Iesato K, Yoshida H, Wakashin Y, Wakashin M, Okuda K. Liver-specific F antigen in the serum of patients with liver diseases and the detection of early stage hepatocellular carcinoma. Clin Chim Acta. 1987;164:127-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 3. | Foster GR, Goldin RD, Oliveira DB. Serum F protein: a new sensitive and specific test of hepatocellular damage. Clin Chim Acta. 1989;184:85-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 13] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 4. | Grewal GK, Oliveira DB. Why serum F protein is a sensitive and specific marker of liver damage in man. Clin Chim Acta. 1990;191:93-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 5. | Teuber SS, Coppel RL, Ansari AA, Leung PS, Neve R, Mackay IR, Gershwin ME. The identification and cloning of the murine genes encoding the liver specific F alloantigens. J Autoimmun. 1991;4:857-870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 6. | Mitchison NA, Simon K. Dominant reduced responsiveness controlled by H-2(Kb)Ab. A new pattern evoked by Thy-1 antigen and F liver antigen. Immunogenetics. 1990;32:104-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 7. | Callaghan N, Majeed T, O'Connell A, Oliveira DB. A comparative study of serum F protein and other liver function tests as an index of hepatocellular damage in epileptic patients. Acta Neurol Scand. 1994;89:237-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 11] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 8. | Cooper CM, Foster GR, Cooper AE, Kenny M, Olivera DB. Serum F-protein concentration following halothane or isoflurane anaesthesia. Clin Chim Acta. 1991;202:73-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 9. | Yang C, Shi YP, Udhayakumar V, Alpers MP, Povoa MM, Hawley WA, Collins WE, Lal AA. Sequence variations in the non-repetitive regions of the liver stage-specific antigen-1 (LSA-1) of Plasmodium falciparum from field isolates. Mol Biochem Parasitol. 1995;71:291-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 10. | Jia C, Zheng S, Zhu Y. Intrathymic inoculation of liver specific antigen alleviates liver transplant rejection. Chin Med Sci J. 2004;19:38-43. [PubMed] |
| 11. | Endo F, Tanaka Y, Tomoeda K, Tanoue A, Tsujimoto G, Nakamura K. Animal models reveal pathophysiologies of tyrosinemias. J Nutr. 2003;133:2063S-2067S. [PubMed] |
| 12. | Liu SY, Xin XT, Cui YM, Yang JS. Purification and clinical application of human liver specific protein. Zhongguo Yixue Jianyan Zazhi. 2001;2:378-381. |
| 13. | Consumer Laboratory Manual of PCR and Gene Clone Regent Box. 2002. Shanghai Shenggong Bio-Engineering Company. . |
| 14. | Forbes GM, Oliveira DB, Hughes RD, O'Grady JG, Calne RY, Williams R. Serum F protein estimation in liver allograft recipients with graft dysfunction. Transplantation. 1989;48:995-997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 15. | Jia CK, Zheng SS, Wan YL, Zhao ZC, Shi SH, Xie HY, Jiang GP, Song PH. The alloimmunogenicity of liver specific antigen. Zhongguo Qiguan Yizhi Zazhi. 2002;6:353-356. |
| 16. | Oliveira DB, Vindlacheruvu S. The phylogenetic distribution of the liver protein F antigen. Comp Biochem Physiol B. 1987;87:87-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
S- Editor Liu Y L- Editor Wang XL E- Editor Zhou T