Published online Jan 7, 2007. doi: 10.3748/wjg.v13.i1.74
Revised: August 25, 2006
Accepted: September 28, 2006
Published online: January 7, 2007
Hepatocellular carcinoma (HCC) is one of the most common cancers in the world with an annual incidence of more than 500 000 in the year 2000. Its incidence is rising in many countries. Recently, it has been estimated that about 53% of HCC cases in the world are related to hepatitis B virus (HBV). The epidemiological association of HBV with HCC is well established. In recent studies, it was revealed that HBsAg carriers have a 25-37 times increased risk of developing HCC as compared to non-infected people. At present, HBV-associated carcinogenesis can be seen as a multi-factorial process that includes both direct and indirect mechanisms that might act synergistically. The integration of HBV DNA into the host genome occurs at early steps of clonal tumor expansion. The integration has been shown in a number of cases to affect a variety of cancer-related genes and to exert insertional mutagenesis. The permanent liver inflammation, induced by the immune response, resulting in a degeneration and regeneration process confers to the accumulation of critical mutations in the host genome. In addition to this, the regulatory proteins HBx and the PreS2 activators that can be encoded by the integrate exert a tumor promoter-like function resulting in positive selection of cells producing a functional regulatory protein. Gene expression profiling and proteomic techniques may help to characterize the molecular mechanisms driving HBV-associated carcinogenesis, and thus potentially identify new strategies in diagnosis and therapy.
- Citation: Lupberger J, Hildt E. Hepatitis B virus-induced oncogenesis. World J Gastroenterol 2007; 13(1): 74-81
- URL: https://www.wjgnet.com/1007-9327/full/v13/i1/74.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i1.74
With an estimate of more than 500 000 incidences in the year 2000 hepatocellular carcinoma (HCC) is one of the most frequent malignant tumors worldwide and its incidence is rising in many countries[1-4]. Despite being the 5th most frequent cancer in the world, HCC is the third leading cause of cancer death behind lung and stomach cancer. The high mortality associated with HCC is due to its unresponsiveness to treatment in many cases and symptoms of HCC often are recognized lately[5]. When viewed as estimated age-adjusted incidence rates of liver cancer per 100 000 men, the figures ranged as follows: in Asia, from 35.5 in Eastern Asia, 18.3 in South-eastern Asia to 5.6 in Western Asia; in Africa, from 24.2 in Middle Africa, 14.4 in Eastern Africa, 13.5 in Western Africa, 6.2 in Southern Africa to 4.9 in Northern Africa; in Europe, from 9.8 in Southern Europe, 5.8 in Eastern and Western Europe to 2.6 in Northern Europe; and to values of 4.8 in South America; 4.1 in North America; 3.6 in Australia/New Zealand and, finally, 2.1 in central America. In all regions, the rates recorded were two to three times higher in men than in women.
These significant differences in the geographic distribution of HCC incidence have led to identify chronic HBV infection as a leading risk factor for HCC[6-9]. Recently, it has been estimated that about 53% of HCC cases in the world are related to HBV[3]. The lifetime risk to develop a HCC was found to be increased even in patients that have cleared hepatitis B virus surface antigen (HBsAg) or with an occult HBV infection. Further risk factors include chronic HCV infection, exposure to aflatoxin B1, alcohol abuse, obesity and diabetes. Aflatoxin B1 (AFB1) is a fungal metabolite that contaminates the food supply in certain areas of the world. It is produced by Aspergillus flavus and related fungi that grow on improperly stored foods, such as corn, rice and peanuts. AFB1 requires metabolic conversion to its exo-8,9-epoxide in order to damage DNA. Coexistence of these risk factors, such as HBV and HCV infection or HBV infection and aflatoxin B1, increases the relative risk of HCC development[11-13]. While a variety of risk factors have been identified in the last years, here a short review describing the current state of knowledge of the molecular pathogenesis of HBV-associated HCC is given. A focus of this review will be on the role of the HBV-regulatory proteins in this process.
The epidemiological association of HBV with HCC is well established. In recent studies, it was revealed that HBsAg carriers have 25-37 times increased risk of developing HCC as compared to non-infected people[14,15]. Moreover, it was analyzed in more detail whether the viral status of the patients are correlated with the risk of developing HCC. HBV has been designated eight genotypes (A-H) based on genetic divergence. Each genotype has a distinct geographical and ethnic distribution. While genotypes B and C are prevalent in Asia, genotypes A and D occur frequently in Africa, Europe and India. There are conflicting data about the influence of HBV genotypes on HCC development[16-18]. Recent studies from Taiwan provide profound evidence for hepatitis B virus e antigen (HBeAg)-positive patients that HBV genotype C causes a more aggressive disease course as compared to genotype B[19-21]. On the other hand, there are reports from Taiwan describing that more than 50% of the HBV-related HCC patients are infected with genotype B. A study on Taiwanese pediatric patients with chronic HBV infection, who were followed for 15 years, showed that genotype B was identified in 74% of the children with HBV-associated HCC[22]. A further interesting observation is the prevalence of the T1762/A1764 mutation in the basal core promoter region which increases with the progression of liver disease. Since this mutation seems to be associated with HCC development, it might represent a helpful prognostic biomarker[23,24].
The risk of HCC seems to be elevated with increasing HBV viral load[25]. Therefore, it is important to consider that most epidemiological analyses were based only on HBsAg positivity. A recent study revealed that the relative risk of HCC was increased by 6-fold among patients who were positive for both HBsAg and HBeAg, compared to those who were positive for HBsAg alone[15]. Based on this, it can be concluded that HBeAg could be an additional useful marker for risk of developing HCC, since HBeAg reflects productive HBV replication.
Integration is not essential for the viral replication but it allows persistence of the viral genome. Almost all of the HBV-associated HCCs harbor chromosomally integrated HBV DNA[26-28]. In many cases, these integrated viral genomes are characterized by rearrangements and/or partial deletions. HBV integration can induce deletions in the host chromosome at the integration site[29]. Based on these observations, it was tempting to speculate that the integration event per se causes a deregulation of key regulators of cell cycle control. This cis-hypothesis (place of integration = place of function) seems to be supported by the woodchuck hepatitis B virus (WHV)-related HCC. Here, insertions of WHV-DNA into the c-myc or, preferentially the N-myc2 gene, have been frequently detected[30-34]. However, in case of the HBV-associated HCC, site-specific integration of the HBV genome or integration of the HBV genome into known oncogenes seems to be a rare event. Interesting examples are the integration of HBV DNA in a cyclin A gene[35], in the retinoic acid receptor beta gene, in the mevalonate kinase gene or in the sarco/endoplasmic reticulum calcium ATPase1 gene[28,36].
It was recently confirmed, using a PCR-based approach, that HBV insertion into cellular genes is a frequent event that occurs early during HBV infection even after acute self-limiting hepatitis[37] and that integration can occur in genes regulating cellular signal transduction cascades, proliferation control and cell viability. Recently, hTERT (human telomerase reverse transcriptase) that is part of the telomerase ribonuclear protein complex was found to be targeted in different HBV-associated HCCs[28,38,39].
In light of these recent data, it will be an important issue to reconsider the role of the integration process for HBV-associated carcinogenesis. A helpful tool will be combining the analysis of putative HBV-specific integration sites with functional genomics of HBV-associated HCCs[40].
In most integrated subviral HBV genomes, the open reading frame for HBx or PreS2 regulatory protein is conserved and can be transcribed[41]. The HBx gene is conserved among all mammalian hepadnaviruses. HBx is a small polypeptide (17 kDa) that is produced at very low levels during chronic and acute hepatitis. Recently, a HBx-like regulatory protein was identified for duck hepatitis B virus (DHBV)[42]. Since the time when HBx initially was described to act as a transcriptional activator[43,44], a variety of functions have been ascribed to the still enigmatic HBx[45,46]. While the X protein is essential for viral replication in case of WHV[47], there are conflicting results about the relevance of HBx for the viral life cycle in case of HBV. There are reports describing that expression of the viral genome occurs independently from HBx functionality[48-50]; other papers describe a relevance of HBx for HBV replication[51]. In transgenic mouse models harboring an overgenomic HBV integrate, it could be observed that HBV replication does not depend on the presence of a functional HBx[52]. Comparable results were obtained in cell culture models based on huh-7 cells[48,50] while in case of HepG2 cells a reduction in HBx-deficient HBV genomes could be observed[53,54]. Moreover, infection experiments of primary tupaia hepatocytes revealed that HBx-deficient HBV particles are infectious (J. Köck, personal communication).
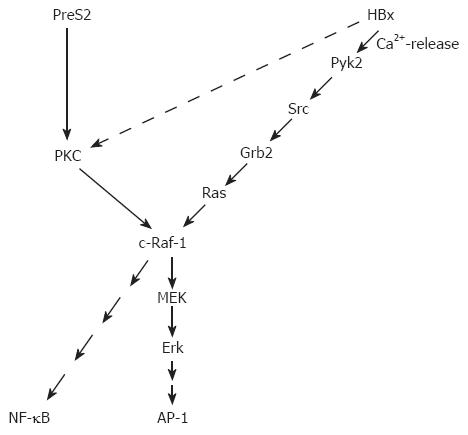
HBx activates a broad variety of different promoter elements. Based on the pleiotropic nature of the HBx-dependent transcriptional regulation, it was concluded that HBx interferes with signaling cascades upstream from the transcription complex. These signaling cascades trigger activation of transcription factors like AP-1 (activator protein-1), NF-κB (nuclear factor kappa B), SP1, and oct-1[46,55]. HBx affects the expression of a variety of genes that are involved in the control of the cell cycle, proliferation or apoptosis. From the beginning, HBx was considered as a crucial viral protein for the process of HBV-associated carcinogenesis[45,56-58] and this might have affected the focus of HBx research. In light of the putative role of HBx for viral carcinogenesis, the major focus of many research projects has been and is the interference of HBx with signal transduction cascades that affect the control of the cell cycle, proliferation or apoptosis. However, one should consider that selective over-expression of HBx reflects a situation that is different from the situation in an infected cell expressing the complete HBV genome. For example, it is well established that HBx is able to promote cellular proliferation[59]. On the other hand, it was shown that expression of the complete HBV genome that harbors the HBx and the PreS2 regulatory protein inhibits cell cycle progression[60] (Figure 1).
The analysis of HBx/protein kinase C (PKC) interaction is such an example for many reports analyzing the interference of HBx with signaling cascades and correlating this with a putative role of HBx for HBV-associated carcinogenesis. There are conflicting reports about the interference of HBx with PKC signaling. On the one hand, there are reports describing an HBx-dependent activation of PKC, mediated by an elevated DAG level in HBx-producing cells. In these studies, PKC is considered as an essential factor for the HBx-dependent activation of NF-kB or AP-1[61,62]. Other reports provide evidence that HBx neither affects activity of PKC nor that PKC is essential for HBx-dependent transcriptional activation[63-65]. The interesting aspect of an HBx-dependent activation of PKC is that a conclusive model for the role of HBx in the process of HBV-dependent carcinogenesis can be deduced. According to the two-step model of carcinogenesis[66] (initiation and promotion), the HBx-dependent activation of PKC could exert a tumor promoter-like function[61]. Independent from the point whether or not the HBx-dependent activation of PKC exerts a tumor promoter-like function, there is profound experimental evidence from experiments with transgenic mice that HBx indeed could exert a tumor promoter-like function. Irradiation of HBx transgenics or exposure of these transgenics to mutagens (diethylnitrosamine) caused a significant increase in the amount of pre-neoplastic lesions as compared to the wild-type control animals[67,68]. Apart from this, a variety of HBx transgenic mouse models were established, but only in one model system so far direct formation of liver cancer could be observed[69].
A tumor promoter-like function of HBx does not necessarily require an activation of PKC. Other pathways as the activation of c-Raf-1-MEK/MAP2 (mitogen-activated protein kinase 2) kinase cascade could fulfill this function as well. Starting with the finding that HBx increases the Ras/GTP complex formation and thereby activates the c-Raf-1 signal transduction cascade[70], more and more data were collected elucidating the interference of HBx with signaling cascades upstream of Ras. One of the next steps was the observation that Src is activated in HBx-producing cells[71,72], followed by the observation that HBx is able to activate the cytosolic Ca2+-dependent praline-rich tyrosine kinase 2 (Pyk)[54]. Pyk is able to activate Src. A recent report describes an HBx-dependent activation of FAK (focal adhesion kinase), a well known regulator of Src kinases[53]. The activation of these signaling cascades requires the presence of HBx in an extranuclear compartment. On the other hand, there is evidence that a fraction of HBx is localized within the nucleus. The subcellular distribution of HBx is still a matter of debate. There are reports providing evidence that HBx is localized in the cytoplasm as well as in the nucleus[49,73]. The different localizations are associated with different functions. HBx localized in the cytoplasm is able to modulate intracellular signal transduction cascades as described above. Moreover, an association of HBx with the outer membrane of mitochondria that induces oxidative stress was described[74-76]. HBx localized in the nucleus is suggested to interfere directly with transcription factors or to exert a transcription factor-like function. A direct interaction with CREB (cAMP responsive element–binding protein) and ATF-2 (activating transcription factor 2) resulting in their increased DNA binding affinity[77] was reported as well as an interaction with RNA polymerase II in the transcription complex[78,79].
In addition to the interaction of HBx with the transcription machinery, there is evidence that HBx interferes at multiple steps with DNA repair and so confers to an increase of critical mutations. HBx was found to bind to DDB1[80,81], a subunit of the damaged DNA binding protein that is bound to damaged DNA, the first step in nucleotide excision repair. In cell culture experiments indeed the expression of HBx significantly inhibited the ability of cells to repair damaged DNA. Therefore, it was tempting to speculate that HBx could confer by this to an increase in the amount of critical mutations in the host genome[80]. However, analysis of mutation frequency in HBx transgenic mice did not corroborate this hypothesis[82]. Other reports focus on the interaction of HBx with p53. On the one hand, it has been shown an indirect inhibition of p53 by HBx: HBx causes a transcriptional repression of the human p53 gene[83]. On the other hand, there is evidence for the capacity of HBx to bind to p53[84,85]. However, if the intracellular amounts of HBx and p53 are considered, there exists a tremendous excess of p53 as compared to HBx in the hepatocytes. The physiological significance of the direct p53/HBx interaction remains questionable.
Apart from the HBx-regulatory protein, the HBV genome encodes a second family of regulatory proteins: the PreS2 activators. Based on a subcloned HBV integrate of the human hepatoma cell line huH4[86] and of an integrate isolated from an HBV-associated HCC[87], preS/S genes that were truncated at the 3´ end were identified[41]. These preS/St genes encoded for C-terminally truncated surface proteins (MHBst) that display a regulatory protein function. Initial analysis revealed that generation of the regulatory protein function requires at least deletion of the last transmembrane region in the S-domain (transmembrane region 3)[88-91]. This results in C-terminally truncated MHBs molecules that are endoplasmic reticulum (ER)-membrane associated by the remaining transmembrane regionsIand II of the S domain[92,93]. A prototype of the ER-membrane-associated MHBst activator is encoded by the integrate isolated from the human hepatoma cell line huH4[86]. This integrate is truncated at ntHBV 221 of the HBV genome resulting in a C-terminally truncated MHBs protein at amino acid (aa) 76 (MHBst76). A detailed analysis revealed that a variety of differences exist between the structural protein MHBs and its C-terminally truncated variant MHBst. In contrast to the structural protein MHBs and the regulatory variants, MHBst are not secreted and lack the glycosylation at asparagine (asn) 4 of the PreS2 domain[92]. The intracellular retention of ER-membrane-associated MHBst proteins gave raise to the hypothesis that the observed activator function is due to ER stress, induced by intracellular retention and subsequent accumulation in the ER[93-96]. More detailed analysis revealed, however, that the structural protein MHBs and the regulatory protein MHBst differ in the topology of the PreS2 domain[97]. In case of the structural protein, the PreS2 domain faces the lumen of the endoplasmic reticulum and in accordance with this glycosylation at asn 4 can occur. In case of the activator protein MHBst, the PreS2 domain directs into the cytoplasm. This explains the lack of N-glycosylation at asn 4. The PreS2 domain facing the cytoplasm interacts with cytosolic binding partners, thereby triggering intracellular signal transduction cascades. In accordance with this, a minimal PreS2 activator was identified lacking any membrane insertion domain (MHBst55)[88,67,98]. This minimal activator encompasses the complete PreS2 domain and is localized within the cytoplasm. Since the PreS2 domain is sufficient to exert the regulatory protein function, this class of regulatory proteins was designated PreS2 activator. The family of PreS2- regulatory proteins encompasses the membrane-associated regulatory proteins, such as MHBst76 or MHBst167, and the non-membrane-associated short proteins, such as PreS2 domain (MHBst55). There is no functional difference between the ER and the cytoplasmatically localized Pres2 activators clearly arguing against the ER-overload hypothesis[97].
The PreS1-PreS2 domain of the large hepatitis B virus surface protein (LHBs) displays a dual membrane topology[99-101]. In one fraction of LHBs, the first transmembrane region that is located at the beginning of the S-domain (aa 8-21) is used: in this case, the PreS1-PreS2 domain of LHBs faces the lumen of the endoplasmic reticulum. In case of the other fraction, this transmembrane region is not used, resulting in a PreS1-PreS2 domain that directs into the cytoplasm. As described above, the cytoplasmic orientation of the PreS2 domain in case of the MHBst proteins is causative for their regulatory protein function. In accordance with this, LHBs displays a regulatory protein function[102] and belongs to the family of PreS2 activator proteins.
The PreS2 activators bind PKC-α in the cytoplasm. This interaction with PKC results in a DAG (1, 2, sn diacylglycerol)-independent activation of PKC and phosphorylation of the PreS2 domain. The activation of PKC is transduced by the c-Raf-1/MEK/ERK (extracellular signal-regulated kinase) signal transduction cascade[63]. This signal transduction cascade can exert a tumor promoter-like function according to the classical two-step model of carcinogenesis[66]. Indeed, transgenic mice expressing the PreS2 activator MHBst76 develop liver tumors at an age above 10 mo. Although the MHBst76 protein is produced in very small but clearly detectable amounts in the MHBst76 transgenic mice, a permanent activation of the Raf-1/MEK/ERK signal transduction cascade can be observed, resulting in an increased proliferation rate of the hepatocytes. The fact that MHBst76 is produced in very small amounts ensures that the observed effects are not due to any overload-associated effects. The tumor formation in these mice can be explained by the permanent activation signal transduction cascades that exert a tumor promoter-like function[63]. Since tumor formation is observed in older animals, it can be assumed that during the aging process critical mutations are accumulated (initiation) and then the tumor promoter function positively selects these cells.
In case of the LHBs-transgenics, tumor formation can be observed as well[103]. In these mice, a very strong overproduction of the LHBs protein occurs, resulting in an intracellular accumulation of the protein and subsequent formation of ground glass hepatocytes. This permanent accumulation results in a situation comparable to a storage disease. Tumor formation in these transgenics was explained by the resulting permanent inflammation[103-105]. In light of the observation that LHBs can act as a regulatory protein, however, the regulatory protein function that is immanent to LHBs should be considered as an additional factor conferring to tumor formation in these mice. The overload-associated stress and inflammation results in the formation of critical mutations (initiation) and the permanent activation of the PKC/Raf/MEK/ERK signal transduction cascade which exerts a tumor promoter-like function.
A major factor in the process of HBV-associated HCC development is the immune system[104,106,107]. The relevance of a chronic, virus-specific immune response for development of HBV-associated carcinoma was shown in an elegant experiment from F. Chisari`s laboratory[108]. Transgenic mice that produce non-cytopathic amounts of HBsAg were used. In these mice, immunologic tolerance against the transgene product can be observed. In accordance with this, no evidence of the liver disease was observed. These mice were subjected to thymectomy and lethally irradiated. One group was reconstituted with the bone marrow and spleen cells derived from non-transgenic littermates that were vaccinated with a recombinant HBsAg encoding vaccinia virus resulting in HBsAg-specific cytotoxic T lymphocytes (CTLs) and antibodies. The other group was reconstituted with the bone marrow and spleen cells derived from transgenic donors that were immunologically tolerant.
In this animal model, the development of hepatitis and later of chronic hepatitis and finally HCC development could be exclusively observed in the mice that were reconstituted with the bone marrow and spleen cells derived from the vaccinated non-transgenic animals, but not in the control groups. Based on this, it was concluded that the immune system-mediated chronic inflammation of the liver, continuous cell death and subsequent cell proliferation might increase the frequency of genetic alterations and the risk of cancer[104,109-111]. This scenario is not exclusively restricted to HBV. Chronic inflammation, degeneration and regeneration are common to a variety of human liver diseases, such as glycogen storage disease or alcoholism or HCV infection, that can finally result in liver carcinoma development[5]. This means that an ineffective immune response can be the principal oncogenic factor during a chronic HBV infection in man. In other words, the same T-cell response can have complete different effects: if the T cell response is strong enough, HBV can be eliminated from the liver, if not, a pro-carcinogenic effect can be induced by permanently triggering necro-inflammatory disease without resulting in a final eradication of HBV from the liver. An interesting aspect is that the nucleoside analogue on lamivudine in patients with chronic hepatitis B can induce the recovery of antiviral T cell responses. However, restoration of HBV-specific T cell reactivity is only transient. The transient nature of the immune reconstitution may represent a favorable condition for virus reactivation once lamivudine therapy is withdrawn.
At present, HBV-associated carcinogenesis can be seen as a multi-factorial process that includes both direct and indirect mechanisms that might act synergistically. The integration of HBV DNA into the host genome occurs at early steps of clonal tumor expansion. The integration has been shown in a number of cases to affect a variety of cancer-related genes and to exert insertional mutagenesis. The permanent liver inflammation resulting in a degeneration and regeneration process confers to the accumulation of critical mutations in the host genome. In addition, the regulatory proteins HBx and the PreS2 activators that can be encoded by the integrate can exert a tumor promoter-like function, resulting in positive selection of cells producing a functional regulatory protein.
Based on new technologies, including gene expression profiling and proteomics, it should be possible to further reveal the molecular mechanisms underlying HBV-associated HCC development and to identify novel diagnostic markers as well as therapeutic and preventive targets.
| 1. | El-Serag HB. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004;127:S27-S34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 738] [Cited by in RCA: 729] [Article Influence: 33.1] [Reference Citation Analysis (1)] |
| 2. | Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907-1917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3241] [Cited by in RCA: 3294] [Article Influence: 143.2] [Reference Citation Analysis (1)] |
| 3. | Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37 Suppl 8:S4-S66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1370] [Cited by in RCA: 1373] [Article Influence: 54.9] [Reference Citation Analysis (0)] |
| 4. | Shibuya K, Mathers CD, Boschi-Pinto C, Lopez AD, Murray CJ. Global and regional estimates of cancer mortality and incidence by site: II. Results for the global burden of disease 2000. BMC Cancer. 2002;2:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 282] [Cited by in RCA: 310] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 5. | Blum HE. Hepatocellular carcinoma: therapy and prevention. World J Gastroenterol. 2005;11:7391-7400. [PubMed] |
| 6. | Ahn SH, Park YN, Park JY, Chang HY, Lee JM, Shin JE, Han KH, Park C, Moon YM, Chon CY. Long-term clinical and histological outcomes in patients with spontaneous hepatitis B surface antigen seroclearance. J Hepatol. 2005;42:188-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 161] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 7. | Evans AA, O'Connell AP, Pugh JC, Mason WS, Shen FM, Chen GC, Lin WY, Dia A, M'Boup S, Dramé B. Geographic variation in viral load among hepatitis B carriers with differing risks of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 1998;7:559-565. [PubMed] |
| 8. | Montalto G, Cervello M, Giannitrapani L, Dantona F, Terranova A, Castagnetta LA. Epidemiology, risk factors, and natural history of hepatocellular carcinoma. Ann N Y Acad Sci. 2002;963:13-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 206] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 9. | Pollicino T, Squadrito G, Cerenzia G, Cacciola I, Raffa G, Craxi A, Farinati F, Missale G, Smedile A, Tiribelli C. Hepatitis B virus maintains its pro-oncogenic properties in the case of occult HBV infection. Gastroenterology. 2004;126:102-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 325] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 10. | Block TM, Mehta AS, Fimmel CJ, Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22:5093-5107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 375] [Article Influence: 16.3] [Reference Citation Analysis (1)] |
| 11. | Kew MC. Synergistic interaction between aflatoxin B1 and hepatitis B virus in hepatocarcinogenesis. Liver Int. 2003;23:405-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 167] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 12. | Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC. TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Sci Publ. 2004;247-270. [PubMed] |
| 13. | Velázquez RF, Rodríguez M, Navascués CA, Linares A, Pérez R, Sotorríos NG, Martínez I, Rodrigo L. Prospective analysis of risk factors for hepatocellular carcinoma in patients with liver cirrhosis. Hepatology. 2003;37:520-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 307] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 14. | Hassan MM, Hwang LY, Hatten CJ, Swaim M, Li D, Abbruzzese JL, Beasley P, Patt YZ. Risk factors for hepatocellular carcinoma: synergism of alcohol with viral hepatitis and diabetes mellitus. Hepatology. 2002;36:1206-1213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 539] [Article Influence: 22.5] [Reference Citation Analysis (1)] |
| 15. | Yang HI, Lu SN, Liaw YF, You SL, Sun CA, Wang LY, Hsiao CK, Chen PJ, Chen DS, Chen CJ. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med. 2002;347:168-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 924] [Cited by in RCA: 917] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 16. | Kao JH. Hepatitis B viral genotypes: clinical relevance and molecular characteristics. J Gastroenterol Hepatol. 2002;17:643-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 249] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 17. | Orito E, Mizokami M. Hepatitis B virus genotypes and hepatocellular carcinoma in Japan. Intervirology. 2003;46:408-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Sugauchi F, Kumada H, Acharya SA, Shrestha SM, Gamutan MT, Khan M, Gish RG, Tanaka Y, Kato T, Orito E. Epidemiological and sequence differences between two subtypes (Ae and Aa) of hepatitis B virus genotype A. J Gen Virol. 2004;85:811-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 116] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 19. | Chen CH, Lee CM, Lu SN, Changchien CS, Eng HL, Huang CM, Wang JH, Hung CH, Hu TH. Clinical significance of hepatitis B virus (HBV) genotypes and precore and core promoter mutations affecting HBV e antigen expression in Taiwan. J Clin Microbiol. 2005;43:6000-6006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 82] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 20. | Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B virus genotypes and spontaneous hepatitis B e antigen seroconversion in Taiwanese hepatitis B carriers. J Med Virol. 2004;72:363-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 140] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 21. | Yu MW, Yeh SH, Chen PJ, Liaw YF, Lin CL, Liu CJ, Shih WL, Kao JH, Chen DS, Chen CJ. Hepatitis B virus genotype and DNA level and hepatocellular carcinoma: a prospective study in men. J Natl Cancer Inst. 2005;97:265-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 417] [Cited by in RCA: 423] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 22. | Ni YH, Chang MH, Wang KJ, Hsu HY, Chen HL, Kao JH, Yeh SH, Jeng YM, Tsai KS, Chen DS. Clinical relevance of hepatitis B virus genotype in children with chronic infection and hepatocellular carcinoma. Gastroenterology. 2004;127:1733-1738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 163] [Article Influence: 7.4] [Reference Citation Analysis (1)] |
| 23. | Kuang SY, Jackson PE, Wang JB, Lu PX, Muñoz A, Qian GS, Kensler TW, Groopman JD. Specific mutations of hepatitis B virus in plasma predict liver cancer development. Proc Natl Acad Sci USA. 2004;101:3575-3580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 124] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 24. | Liu CJ, Chen BF, Chen PJ, Lai MY, Huang WL, Kao JH, Chen DS. Role of hepatitis B viral load and basal core promoter mutation in hepatocellular carcinoma in hepatitis B carriers. J Infect Dis. 2006;193:1258-1265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 141] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 25. | Iloeje UH, Yang HI, Su J, Jen CL, You SL, Chen CJ. Predicting cirrhosis risk based on the level of circulating hepatitis B viral load. Gastroenterology. 2006;130:678-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1164] [Cited by in RCA: 1182] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
| 26. | Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981;2:1129-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1838] [Cited by in RCA: 1761] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 27. | Brechot C, Pourcel C, Louise A, Rain B, Tiollais P. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature. 1980;286:533-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 423] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 28. | Paterlini-Bréchot P, Saigo K, Murakami Y, Chami M, Gozuacik D, Mugnier C, Lagorce D, Bréchot C. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene. 2003;22:3911-3916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 252] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 29. | Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1097] [Cited by in RCA: 1103] [Article Influence: 46.0] [Reference Citation Analysis (1)] |
| 30. | Bruni R, D'Ugo E, Villano U, Fourel G, Buendia MA, Rapicetta M. The win locus involved in activation of the distal N-myc2 gene upon WHV integration in woodchuck liver tumors harbors S/MAR elements. Virology. 2004;329:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 31. | Buendia MA. Hepatitis B viruses and cancerogenesis. Biomed Pharmacother. 1998;52:34-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 32. | Fourel G, Trepo C, Bougueleret L, Henglein B, Ponzetto A, Tiollais P, Buendia MA. Frequent activation of N-myc genes by hepadnavirus insertion in woodchuck liver tumours. Nature. 1990;347:294-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 184] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 33. | Hsu T, Möröy T, Etiemble J, Louise A, Trépo C, Tiollais P, Buendia MA. Activation of c-myc by woodchuck hepatitis virus insertion in hepatocellular carcinoma. Cell. 1988;55:627-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 135] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 34. | Jacob JR, Sterczer A, Toshkov IA, Yeager AE, Korba BE, Cote PJ, Buendia MA, Gerin JL, Tennant BC. Integration of woodchuck hepatitis and N-myc rearrangement determine size and histologic grade of hepatic tumors. Hepatology. 2004;39:1008-1016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 35. | Wang J, Chenivesse X, Henglein B, Bréchot C. Hepatitis B virus integration in a cyclin A gene in a hepatocellular carcinoma. Nature. 1990;343:555-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 492] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 36. | Bréchot C, Gozuacik D, Murakami Y, Paterlini-Bréchot P. Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). Semin Cancer Biol. 2000;10:211-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 226] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 37. | Murakami Y, Saigo K, Takashima H, Minami M, Okanoue T, Bréchot C, Paterlini-Bréchot P. Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut. 2005;54:1162-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 246] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 38. | Horikawa I, Barrett JC. cis-Activation of the human telomerase gene (hTERT) by the hepatitis B virus genome. J Natl Cancer Inst. 2001;93:1171-1173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 70] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 39. | Hytiroglou P, Theise ND. Telomerase activation in human hepatocarcinogenesis. Am J Gastroenterol. 2006;101:839-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 40. | Thorgeirsson SS, Lee JS, Grisham JW. Functional genomics of hepatocellular carcinoma. Hepatology. 2006;43:S145-S150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 101] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 41. | Schlüter V, Meyer M, Hofschneider PH, Koshy R, Caselmann WH. Integrated hepatitis B virus X and 3' truncated preS/S sequences derived from human hepatomas encode functionally active transactivators. Oncogene. 1994;9:3335-3344. [PubMed] |
| 42. | Chang SF, Netter HJ, Hildt E, Schuster R, Schaefer S, Hsu YC, Rang A, Will H. Duck hepatitis B virus expresses a regulatory HBx-like protein from a hidden open reading frame. J Virol. 2001;75:161-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 43. | Twu JS, Schloemer RH. Transcriptional trans-activating function of hepatitis B virus. J Virol. 1987;61:3448-3453. [PubMed] |
| 44. | Wollersheim M, Debelka U, Hofschneider PH. A transactivating function encoded in the hepatitis B virus X gene is conserved in the integrated state. Oncogene. 1988;3:545-552. [PubMed] |
| 45. | Cougot D, Neuveut C, Buendia MA. HBV induced carcinogenesis. J Clin Virol. 2005;34 Suppl 1:S75-S78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 133] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 46. | Zhang X, Zhang H, Ye L. Effects of hepatitis B virus X protein on the development of liver cancer. J Lab Clin Med. 2006;147:58-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 156] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 47. | Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026-2030. [PubMed] |
| 48. | Blum HE, Zhang ZS, Galun E, von Weizsäcker F, Garner B, Liang TJ, Wands JR. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J Virol. 1992;66:1223-1227. [PubMed] |
| 49. | Hafner A, Brandenburg B, Hildt E. Reconstitution of gene expression from a regulatory-protein-deficient hepatitis B virus genome by cell-permeable HBx protein. EMBO Rep. 2003;4:767-773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 50. | Stöckl L, Berting A, Malkowski B, Foerste R, Hofschneider PH, Hildt E. Integrity of c-Raf-1/MEK signal transduction cascade is essential for hepatitis B virus gene expression. Oncogene. 2003;22:2604-2610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 51. | Melegari M, Wolf SK, Schneider RJ. Hepatitis B virus DNA replication is coordinated by core protein serine phosphorylation and HBx expression. J Virol. 2005;79:9810-9820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 113] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 52. | Reifenberg K, Nusser P, Löhler J, Spindler G, Kuhn C, von Weizsäcker F, Köck J. Virus replication and virion export in X-deficient hepatitis B virus transgenic mice. J Gen Virol. 2002;83:991-996. [PubMed] |
| 53. | Bouchard MJ, Wang L, Schneider RJ. Activation of focal adhesion kinase by hepatitis B virus HBx protein: multiple functions in viral replication. J Virol. 2006;80:4406-4414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 86] [Article Influence: 4.3] [Reference Citation Analysis (1)] |
| 54. | Bouchard MJ, Wang LH, Schneider RJ. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294:2376-2378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 327] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 55. | Waris G, Siddiqui A. Regulatory mechanisms of viral hepatitis B and C. J Biosci. 2003;28:311-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 60] [Article Influence: 2.6] [Reference Citation Analysis (1)] |
| 56. | Chan HL, Sung JJ. Hepatocellular carcinoma and hepatitis B virus. Semin Liver Dis. 2006;26:153-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 177] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 57. | Koike K, Tsutsumi T, Fujie H, Shintani Y, Kyoji M. Molecular mechanism of viral hepatocarcinogenesis. Oncology. 2002;62 Suppl 1:29-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 93] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 58. | Staib F, Hussain SP, Hofseth LJ, Wang XW, Harris CC. TP53 and liver carcinogenesis. Hum Mutat. 2003;21:201-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 175] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 59. | Madden CR, Slagle BL. Stimulation of cellular proliferation by hepatitis B virus X protein. Dis Markers. 2001;17:153-157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 60. | Friedrich B, Wollersheim M, Brandenburg B, Foerste R, Will H, Hildt E. Induction of anti-proliferative mechanisms in hepatitis B virus producing cells. J Hepatol. 2005;43:696-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 61. | Kekulé AS, Lauer U, Weiss L, Luber B, Hofschneider PH. Hepatitis B virus transactivator HBx uses a tumour promoter signalling pathway. Nature. 1993;361:742-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 277] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 62. | Luber B, Lauer U, Weiss L, Höhne M, Hofschneider PH, Kekulé AS. The hepatitis B virus transactivator HBx causes elevation of diacylglycerol and activation of protein kinase C. Res Virol. 1993;144:311-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 63. | Hildt E, Munz B, Saher G, Reifenberg K, Hofschneider PH. The PreS2 activator MHBs(t) of hepatitis B virus activates c-raf-1/Erk2 signaling in transgenic mice. EMBO J. 2002;21:525-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 143] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 64. | Lucito R, Schneider RJ. Hepatitis B virus X protein activates transcription factor NF-kappa B without a requirement for protein kinase C. J Virol. 1992;66:983-991. [PubMed] |
| 65. | Murakami S, Cheong J, Ohno S, Matsushima K, Kaneko S. Transactivation of human hepatitis B virus X protein, HBx, operates through a mechanism distinct from protein kinase C and okadaic acid activation pathways. Virology. 1994;199:243-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 66. | Boutwell RK. The function and mechanism of promoters of carcinogenesis. CRC Crit Rev Toxicol. 1974;2:419-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 493] [Cited by in RCA: 449] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 67. | Madden CR, Finegold MJ, Slagle BL. Hepatitis B virus X protein acts as a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J Virol. 2001;75:3851-3858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 106] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 68. | Zhu H, Wang Y, Chen J, Cheng G, Xue J. Transgenic mice expressing hepatitis B virus X protein are more susceptible to carcinogen induced hepatocarcinogenesis. Exp Mol Pathol. 2004;76:44-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 41] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 69. | Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 880] [Cited by in RCA: 875] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 70. | Benn J, Schneider RJ. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc Natl Acad Sci USA. 1994;91:10350-10354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 335] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 71. | Klein NP, Bouchard MJ, Wang LH, Kobarg C, Schneider RJ. Src kinases involved in hepatitis B virus replication. EMBO J. 1999;18:5019-5027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 98] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 72. | Klein NP, Schneider RJ. Activation of Src family kinases by hepatitis B virus HBx protein and coupled signaling to Ras. Mol Cell Biol. 1997;17:6427-6436. [PubMed] |
| 73. | Sirma H, Weil R, Rosmorduc O, Urban S, Israël A, Kremsdorf D, Bréchot C. Cytosol is the prime compartment of hepatitis B virus X protein where it colocalizes with the proteasome. Oncogene. 1998;16:2051-2063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 83] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 74. | Huh KW, Siddiqui A. Characterization of the mitochondrial association of hepatitis B virus X protein, HBx. Mitochondrion. 2002;1:349-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 75. | Lee YI, Hwang JM, Im JH, Lee YI, Kim NS, Kim DG, Yu DY, Moon HB, Park SK. Human hepatitis B virus-X protein alters mitochondrial function and physiology in human liver cells. J Biol Chem. 2004;279:15460-15471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 150] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 76. | Waris G, Huh KW, Siddiqui A. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-kappa B via oxidative stress. Mol Cell Biol. 2001;21:7721-7730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 283] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 77. | Maguire HF, Hoeffler JP, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;252:842-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 371] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 78. | Haviv I, Shamay M, Doitsh G, Shaul Y. Hepatitis B virus pX targets TFIIB in transcription coactivation. Mol Cell Biol. 1998;18:1562-1569. [PubMed] |
| 79. | Haviv I, Vaizel D, Shaul Y. pX, the HBV-encoded coactivator, interacts with components of the transcription machinery and stimulates transcription in a TAF-independent manner. EMBO J. 1996;15:3413-3420. [PubMed] |
| 80. | Becker SA, Lee TH, Butel JS, Slagle BL. Hepatitis B virus X protein interferes with cellular DNA repair. J Virol. 1998;72:266-272. [PubMed] |
| 81. | Wentz MJ, Becker SA, Slagle BL. Dissociation of DDB1-binding and transactivation properties of the hepatitis B virus X protein. Virus Res. 2000;68:87-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 82. | Madden CR, Finegold MJ, Slagle BL. Expression of hepatitis B virus X protein does not alter the accumulation of spontaneous mutations in transgenic mice. J Virol. 2000;74:5266-5272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 83. | Lee SG, Rho HM. Transcriptional repression of the human p53 gene by hepatitis B viral X protein. Oncogene. 2000;19:468-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 97] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 84. | Elmore LW, Hancock AR, Chang SF, Wang XW, Chang S, Callahan CP, Geller DA, Will H, Harris CC. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc Natl Acad Sci USA. 1997;94:14707-14712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 255] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 85. | Ueda H, Ullrich SJ, Gangemi JD, Kappel CA, Ngo L, Feitelson MA, Jay G. Functional inactivation but not structural mutation of p53 causes liver cancer. Nat Genet. 1995;9:41-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 255] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 86. | Kekulé AS, Lauer U, Meyer M, Caselmann WH, Hofschneider PH, Koshy R. The preS2/S region of integrated hepatitis B virus DNA encodes a transcriptional transactivator. Nature. 1990;343:457-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 206] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 87. | Caselmann WH, Meyer M, Kekulé AS, Lauer U, Hofschneider PH, Koshy R. A trans-activator function is generated by integration of hepatitis B virus preS/S sequences in human hepatocellular carcinoma DNA. Proc Natl Acad Sci USA. 1990;87:2970-2974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 99] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 88. | Lauer U, Weiss L, Hofschneider PH, Kekulé AS. The hepatitis B virus pre-S/S(t) transactivator is generated by 3' truncations within a defined region of the S gene. J Virol. 1992;66:5284-5289. [PubMed] |
| 89. | Levrero M, Balsano C, Avantaggiati ML, Natoli G, Chirillo P, De Marzio E, Collepardo D, Costanzo A. Characterization of the hepatitis B virus transactivators: a possible direct role of the virus in the development of hepatocellular carcinoma. J Surg Oncol Suppl. 1993;3:34-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 90. | Natoli G, Avantaggiati ML, Balsano C, De Marzio E, Collepardo D, Elfassi E, Levrero M. Characterization of the hepatitis B virus preS/S region encoded transcriptional transactivator. Virology. 1992;187:663-670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 91. | Natoli G, Balsano C, Avantaggiati ML, De Marzio E, Artini M, Collepardo D, Elfassi E, Levrero M. Truncated pre-S/S proteins transactivate multiple target sequences. Arch Virol Suppl. 1992;4:65-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 92. | Hildt E, Urban S, Lauer U, Hofschneider PH, Kekulé AS. ER-localization and functional expression of the HBV transactivator MHBst. Oncogene. 1993;8:3359-3367. [PubMed] |
| 93. | Meyer M, Caselmann WH, Schlüter V, Schreck R, Hofschneider PH, Baeuerle PA. Hepatitis B virus transactivator MHBst: activation of NF-kappa B, selective inhibition by antioxidants and integral membrane localization. EMBO J. 1992;11:2991-3001. [PubMed] |
| 94. | Caselmann WH, Renner M, Schlüter V, Hofschneider PH, Koshy R, Meyer M. The hepatitis B virus MHBst167 protein is a pleiotropic transactivator mediating its effect via ubiquitous cellular transcription factors. J Gen Virol. 1997;78 (Pt 6):1487-1495. [PubMed] |
| 95. | Jüngst C, Cheng B, Gehrke R, Schmitz V, Nischalke HD, Ramakers J, Schramel P, Schirmacher P, Sauerbruch T, Caselmann WH. Oxidative damage is increased in human liver tissue adjacent to hepatocellular carcinoma. Hepatology. 2004;39:1663-1672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 100] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 96. | Xu Z, Jensen G, Yen TS. Activation of hepatitis B virus S promoter by the viral large surface protein via induction of stress in the endoplasmic reticulum. J Virol. 1997;71:7387-7392. [PubMed] |
| 97. | Hildt E, Urban S, Hofschneider PH. Characterization of essential domains for the functionality of the MHBst transcriptional activator and identification of a minimal MHBst activator. Oncogene. 1995;11:2055-2066. [PubMed] |
| 98. | Hildt E, Urban S, Eckerskorn C, Hofschneider PH. Isolation of highly purified, functional carboxy-terminally truncated hepatitis B virus middle surface protein activators from eucaryotic expression systems. Hepatology. 1996;24:502-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 99. | Bruss V, Lu X, Thomssen R, Gerlich WH. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 1994;13:2273-2279. [PubMed] |
| 100. | Ostapchuk P, Hearing P, Ganem D. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 1994;13:1048-1057. [PubMed] |
| 101. | Prange R, Streeck RE. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 1995;14:247-256. [PubMed] |
| 102. | Hildt E, Saher G, Bruss V, Hofschneider PH. The hepatitis B virus large surface protein (LHBs) is a transcriptional activator. Virology. 1996;225:235-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 125] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 103. | Chisari FV, Klopchin K, Moriyama T, Pasquinelli C, Dunsford HA, Sell S, Pinkert CA, Brinster RL, Palmiter RD. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell. 1989;59:1145-1156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 535] [Cited by in RCA: 520] [Article Influence: 14.1] [Reference Citation Analysis (1)] |
| 104. | Chisari FV. Rous-Whipple Award Lecture. Viruses, immunity, and cancer: lessons from hepatitis B. Am J Pathol. 2000;156:1117-1132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 230] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 105. | Dunsford HA, Sell S, Chisari FV. Hepatocarcinogenesis due to chronic liver cell injury in hepatitis B virus transgenic mice. Cancer Res. 1990;50:3400-3407. [PubMed] |
| 106. | Rehermann B. Immune responses in hepatitis B virus infection. Semin Liver Dis. 2003;23:21-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 78] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 107. | Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1202] [Cited by in RCA: 1225] [Article Influence: 58.3] [Reference Citation Analysis (0)] |
| 108. | Nakamoto Y, Guidotti LG, Kuhlen CV, Fowler P, Chisari FV. Immune pathogenesis of hepatocellular carcinoma. J Exp Med. 1998;188:341-350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 283] [Cited by in RCA: 261] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 109. | Chisari FV, Pinkert CA, Milich DR, Filippi P, McLachlan A, Palmiter RD, Brinster RL. A transgenic mouse model of the chronic hepatitis B surface antigen carrier state. Science. 1985;230:1157-1160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 457] [Cited by in RCA: 354] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 110. | Ferrari C, Missale G, Boni C, Urbani S. Immunopathogenesis of hepatitis B. J Hepatol. 2003;39 Suppl 1:S36-S42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 53] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 111. | Visvanathan K, Lewin SR. Immunopathogenesis: role of innate and adaptive immune responses. Semin Liver Dis. 2006;26:104-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
S- Editor Liu Y L- Editor Kumar M E- Editor Ma WH