Published online Feb 21, 2006. doi: 10.3748/wjg.v12.i7.1025
Revised: June 2, 2005
Accepted: June 11, 2005
Published online: February 21, 2006
AIM: To investigate the differently expressed genes in human colorectal adenocarcinoma.
METHODS: The integrated approach for gene expression profiling that couples suppression subtractive hybridization, high-throughput cDNA array, sequencing, bioinformatics analysis, and reverse transcriptase real-time quantitative polymerase chain reaction (PCR) was carried out. A set of cDNA clones including 1260 SSH inserts amplified by PCR was arrayed using robotic printing. The cDNA arrays were hybridized with florescent-labeled probes prepared from RNA of human colorectal adenocarcinoma (HCRAC) and normal colorectal tissues.
RESULTS: A total of 86 genes were identified, 16 unknown genes and 70 known genes. The transcription factor Sox9 influencing cell differentiation was downregulated. At the same time, Heat shock protein 10 KDis downregulated and Calmoulin is up-regulated.
CONCLUSION: Downregulation of heat shock protein 10 KD lost its inhibition of Ras, and then attenuated the Ras GTPase signaling pathway, increased cell proliferation and inhibited cell apoptosis. Down-regulated transcription factor So x 9 influences cell differentiation and cell-specific gene expression. Down-regulated So x 9 also decreases its binding to calmodulin, accumulates calmodulin as receptor-activated kinase and phosphorylase kinase due to the activation of PhK.
- Citation: Chen Y, Zhang YZ, Zhou ZG, Wang G, Yi ZN. Identification of differently expressed genes in human colorectal adenocarcinoma. World J Gastroenterol 2006; 12(7): 1025-1032
- URL: https://www.wjgnet.com/1007-9327/full/v12/i7/1025.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i7.1025
The lifetime risk of colorectal cancer in the general population is about 5-6% and about 30-40% of patients have cancer-related deaths[1]. It is of great importance to elucidate the mechanisms involved in human colorectal gene carcinogenesis at cellular and molecular levels. It has been demonstrated that colorectal tumor undergoes multiple and sequential morphological and molecular changes[2]. In the process of the disease, cell division, differentiation and apoptosis are related to the expression disorder of a group of genes and protein interaction. Oncogenes such as myc, K-ras, src, and erbB2 are activated, while the suppressive cancer genes p53, DCC, APC lose their activation and mismatch repair leads to microsatellite instability[3-9]. However, the exact gene expression profiling for human colorectal adenocarcinoma (HCRAC) is still unclear.
Suppression subtractive hybridization (SSH) techniques based on suppression polymerase chain reaction (PCR) can identify the gene expression profiling[10]. We have described recently the successful use of SSH using HCRAC and normal colorectal tissues[11]. Combined with cDNA microarray techniques, the gene expressing profiling of HCRAC can be obtained with four additional pairs of normal colorectal and HCRAC tissues. In this study, 86 genes were identified. According to the results of sequencing and bioinformatics analyses, three genes were selected, which differed strongly between HCRAC and normal colorectal tissues, and four genes were evaluated in 10 pairs of matched normal colorectal and HCRAC tissues by QRT-PCR technique.
Normal colorectal and HCRAC tissue samples from HCRAC patients in West China Hospital were snap-frozen in liquid nitrogen immediately after surgery and stored at -80 °C. Ten pairs of HCRAC and normal colorectal tissues were used. All the tumor samples containing >50% of tumor cells were confirmed as colorectal adenocarcinoma according to the result of pathologic diagnosis.
Isolation of RNA was performed using the TRIzol method (Invitrogen) according to manufacturer’s instructions. RNA from each sample was assessed by visualization of the 28S/18S ribosomal RNA ratio on 1% agarose gel. RNA yield was determined by measuring the absorbency at 260 nm.
PCR-based cDNA subtraction method and PCR-select subtraction method (Clontech Laboratories) were performed using the SMART PCR cDNA synthesis as described previously[10,11]. Total RNA was isolated from HCRAC and normal rectum tissues, respectively, and reversely transcripted into single-strand cDNAs with reverse transcript enzyme superscript II. Double-strand cDNA was then synthesized and digested with restriction enzyme RsaI, resulting in fragments with a different size of 400-600 bp. HCRAC cDNAs were divided into two groups and ligated to the specific adaptors 1 and 2R, respectively. After HCRAC cDNAs were hybridized with normal rectum cDNA twice for 10 h at 68 °C and the resulting cDNAs were amplified with nested PCR twice, the amplified cDNAs containing enriched differently expressed transcripts were cloned into plasmid vector arms of T/A. The ligated cDNAs were transformed into E. coli strain JM109.
A cDNA gene chip containing 1260 SSH clones was made .The inserted fragments in the pMD-18T (TaKaRa Company) were amplified with PCR using primers from the franking cloning site. Then the amplified PCR products were spotted onto silylated slides (CEL Associates, Houston, TX, USA) using a Cartesian PixSys 7500 motion control robot (Cartesian Technologies, Irvine, CA, USA) fitted with ChipMaker Micro-Spotting Technology (TeleChem International, Sunnyvale, CA, USA). The total RNA from the four pairs of samples taken from grade A patients was used to prepare cDNA fluorescent probes for hybridization to microarray. Probes were prepared by reverse transcription of total RNA in the presence of either Cy-5 or Cy-3 labeled dUTP (Amersham Pharmacia) using superscript II (Gibco-BRL). The two color probes were then mixed, and the denatured probe mixtures were applied onto the pre-hybridized chip under a cover glass. The chips were hybridized at 42 °C for 15-17 h and scanned with a ScanArray 3000 (GSI Lumonics, Billerica, MA, USA) at two wavelengths to detect emission from both Cy3 and Cy5. The acquired images were analyzed using ImaGene 3.0 software (BioDiscovery, Inc., Los Angeles, CA, USA). The intensities of each spot at the two wavelengths represented the quantity of Cy3-dUTP and Cy5-dUTP, respectively, hybridized to each spot. Genes were identified as differently expressed, if the absolute value of the natural logarithm of the ratios was >0.69. To minimize artifacts arising from low expression values, only genes with raw intensity values for both Cy3 and Cy5 >800 counts were chosen for differential analysis. The clustering algorithm separated tumor and normal tissues into different clusters. We used an algorithm based on the hierarchical clustering algorithm to organize the data matrix of the four colorectal adenocarcinoma samples in a binary tree.
Sequencing was carried out for the cDNA clones using the Thermo Sequenase fluorescent labeled primer cycle sequencing (Amersham Pharmacia Biotech, UK). CR -r6d4cts were run on SequaGel (Biozyme, Oldendorf, Germang) and the sequences were analyzed by a LiCOR sequencer (MWG Biotech, Ebersberg, Germany). The qualified expressed sequence tags (ESTs) were referred to the ESTs containing less than 3% ambiguous bases and longer than 100 bp. These ESTs were subjected to BLAST analysis. The EST was considered as part of a known gene, if the homology to the known gene was over 80%. The EST was considered as part of a known EST, if it shared 95% homology with at least 100 bp of human EST. The EST with no match to human EST was considered as a novel EST.
The total RNA from 10 pairs of patients was treated with RNA-free DNAase I (TaKaRa Company) to remove any genomic DNA contamination, then subjected to reverse transcription using M-MuLV reverse transcriptase (Qiagen Company, Germany).
PCR primers were designed using Primer Express 1.5 (PE Company) with the following parameters: length of the amplicon was between 50 and 150 bp, Tm of the primers was between 58 °C and 60 °C, and span of an intron (Table 1). Real time quantitative (RTQ) PCR was performed using Taqman MGB reagent kit (Qiagen Company, Germany) according to the manufacturer′s instructions. PCR was performed on an ABI Prism 700 fluorescent quantitative PCR.
| Gene | Primer | Sequence | Length of primer (bp) | Length of product (bp) | Location of product |
| HSE1 | |||||
| Forward prime | 5’-TGGCAGGACAAGCGTTTAGA-3’ | 20 | 66 | 43–108 | |
| Reverse prime | 5’-CAGCACTCCTTTCAACCAATACTC-3 | 24 | |||
| TaqMan probe | 5’>FAM AGTTTCTTCCACTCTTTG Quencher < 3’ | 18 | 64–81 | ||
| SOX9 | |||||
| Forward prime | 5’-AGCGACGTCATCTCCAACATC-3’ | 21 | 49 | 1235–1 | |
| Reverse prime | 5’-GTTGGGCGGCAGGTACTG-3’ | 18 | 252 | ||
| TaqMan | 5’>FAM CCTTCGATGTCAACGAGT Quencher < 3’ | 18 | |||
| CaM | 3098–3 | ||||
| Forward prime | 5’-GTTGAGCGAGGCAAATGGAT-3’ | 20 | 62 | 161 | |
| Reverse prime | 5’-TCCTTGGCAACAGTGCATCA-3’ | 20 | |||
| TaqMan | 5’>FAM TCGATATTTCAGATGGGC Quencher <3 ’ | 18 | |||
| β-Actin | 1058–1 | ||||
| n | Forward prime | 5’-CTGGCACCCAGCACAATG-3’ | 18 | 93 | 075 |
| Reverse prime | 5’-GGACAGCGAGGCCAGGAT-3’ | 18 | |||
| TaqMan | 5’>FAM ATCATTGCTCCTCCTGAG Quencher < 3’ | 18 | |||
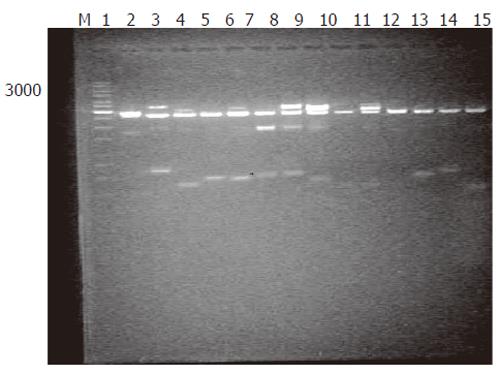
Generation of subtracted cDNA populations: SSH was performed in human rectum adenocarcinoma and normal rectum tissues from one patient. A total of 1 260 clones were obtained by SSH. To evaluate the quality of the libraries, plasmid DNA isolated from 36 clones was digested with restriction enzyme, and agarose gel electrophoresis showed that all plasmids contained 400-600 bp fragments (Figure 1). Sequence analysis of 31 clones demonstrated that 28 fragments were differentially homologous to known genes in the GenBank (acquired gene accession number and dbEST number: Accn BM360856-BM360883).
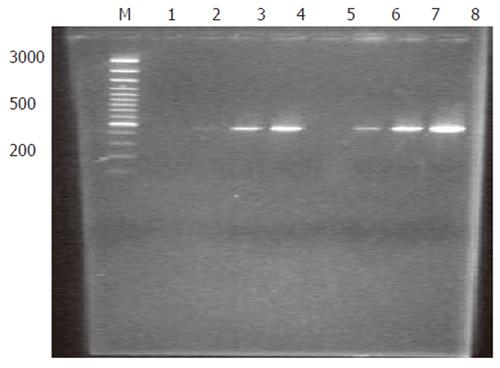
Analysis of subtraction efficiency: To estimate the efficiency of subtraction in subtractive cDNA library, the amount of cDNA for G3PDH and oncogene c-myc was analyzed using subtracted and unsubtracted PCR products as templates. Eighteen, 23, 28 and 33 cycles of PCR reactions were performed in a total reaction volume of 30 µL. The result demonstrated that the non-specifically expressed housekeeping gene G3PDH was greatly decreased in the reaction (Figure 2). In contrast, the specifically expressed oncogene c-myc was greatly increased in the reaction (Figure 3). These results indicated that the present suppression subtractive cDNA library was highly efficient in enriching the highly expressed genes in HCRAC.
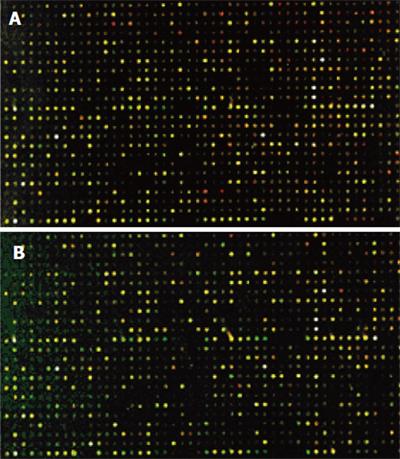
The cDNA inserts of each clone were amplified with PCR and spotted on a microarray using robotic printing. Multiple housekeeping genes and randomly selected cDNAs were also printed on the same array to serve as internal controls. The cDNAs from cancer tissue of four patients were labeled with cyanine5 (Cy5), and the cDNAs from normal colon tissue of four patients were labeled with cyanine3 (Cy3). The microarrays were hybridized with cDNA probes labeled with fluorochrome in the forward direction as group 1(Figure 4A).Then, the cDNAs from cancer tissue of four patients were labeled with Cy3, and the cDNAs from normal colon tissue of four patients were labeled with Cy5. The microarrays were hybridized with cDNA probes labeled with fluorochrome by reverse direction as group 2 (Figure 4B). Red and green fluorescence indicated greater relative expression in the tissues of HCRAC and normal colon, yellow fluorescence indicated equal expression. According to the changes in signal intensity, a total of 143 ESTs were identified. Fifty-four ESTs were differently expressed in group 1, 37 ESTs were upregulated and 17 ESTs were downregulated by forward hybridization. ESTs were differently expressed in group 2 by reverse hybridization, 49 ESTs were upregulated, 40 ESTs were downregulated. The identified 43 ESTs representing 3.41% of total genes examined were differently expressed in the two groups (Table 2).
| Name of gene | Accessionnumber | cDNA | cDNA array ratio | ||
| Group 1 | Group 2 | ||||
| Proto-oncogene and tumor suppressor genes | ITGA6: integrin, alpha 6 | ck570352 | C60 | 2.277 | |
| Ion channels | ATP1B1: ATPase, Na+/K+ transporting, beta 1 | ck570329 | C8 | 0.352 | |
| proteins | polypeptide | ck570304, | a50 | 3.265 | |
| CALM1: calmodulin 1 (phosphorylase kinase, delta) | ck570314 | b19 | 1.856 | 0.344 | |
| Cell cycle proteins | PTMA: prothymosin, alpha | ck570346 | C47 | 2.090 | |
| BTG1: B-cell translocation gene 1, anti-proliferative | ck570300 | a21 | 2.058 | ||
| DKFZp451J0118: taxilin | |||||
| CAPZA1: capping protein (actin filament) muscle | ck570339 | c32 | 0.472 | ||
| Z-line, alpha 1 | ck570343 | c41 | 0.493 | ||
| TBDN100: transcriptional coactivator tubedown-100 | ck570351 | c58 | 2.211 | ||
| Cell frame and movement | K-ALPHA-1: tubulin, alpha, ubiquitous | ck570353 | C61 | 2.281 | |
| TUBB2: tubulin, beta, 2 | ck570366 | C91 | 3.21 | ||
| CAPZA1: capping protein (actin filament) muscle Z-line, alpha 1 | ck570343 | C41 | 0.493 | ||
| Apoptosis related proteins Signal | K-ALPHA-1: tubulin, alpha, ubiquitous | ck570339 | C32 | 0.472 | |
| SRPRB: signal recognition particle receptor, B subunit | ck570312 | b12 | 1.540 | 0.425 | |
| transmitting related proteins | PERQ1: PERQ amino acid rich, with GYF domain 1 | ck570299 | a15 | 0.486 | |
| Synthesis and translation | ZNF403: zinc finger protein 403 | ck570360 | C85 | 3.197 | 3.197 |
| related proteins | SOX9: SRY (sex determining region Y)-box 9 | ck570305, | b2 | 0.426 | 2.344 |
| (campomelic dysplasia, autosomal sex-reversal) | ck570306, | b3 | 0.489 | 1.593 | |
| ck570361 | c86 | 3.391 | |||
| SET: SET translocation (myeloid leukemia-associated) | ck570344 | c43 | 2.447 | ||
| Bit1: Bcl-2 inhibitor of transcription | |||||
| EIF3S6: eukaryotic translation initiation factor 3, subunit | ck570355 | c73 | 2.666 | ||
| 6 48 KD | ck570311 | b11 | 0.660 | 2.091 | |
| EIF4E: eukaryotic translation initiation factor 4E | |||||
| NCL: nucleolin | ck570336 | c16 | 0.392 | ||
| ck570367 | c92 | 4.67 | |||
| Metabolism | RPL7: ribosomal protein L7 | ck570290 | a3 | 0.359 | |
| related proteins | RPS24: ribosomal protein S24 | ck570297 | a13 | 0.457 | |
| RPS25: ribosomal protein S25 | ck570349 | c55 | 2.202 | ||
| COX7C: cytochrome c oxidase subunit VIIc | ck570315 | b20 | 1.860 | 0.580 | |
| FABP1: fatty acid binding protein 1, liver | ck570324 | b39 | 2.807 | 0.547 | |
| SAT: spermidine/spermine N1-acetyltransferase | ck570335 | c13 | 0.375 | ||
| SCD: stearoyl-CoA desaturase (delta-9-desaturase) | ck570357 | c81 | 3.098 | ||
| PAFAH1B2: platelet-activating factor acetylhydrolase, isoform Ib, beta subunit 30 KD protein 1 | ck570292 | a7 | 0.421 | ||
| Immunity and stimulation related proteins | HSPE1: heat shock 10 protein 1 (chaperonin 10) | ck570307, | b6 | 0.562 | 3.191 |
| ck570308, | b7 | 0.567 | 2.686 | ||
| ck570310, | b9 | 0.649 | 2.532 | ||
| ck570347 | c52 | 2.132 | |||
| HSPD1: heat shock 60 KD protein 1 (chaperonin) | ck570356 | c77 | 2.786 | ||
| IGHG1: immunoglobulin heavy constant gamma 1 | ck570363 | c90 | 4.667 | ||
| (G1m marker) | ck570321 | b5-1 | 0.493 | 2.830 | |
| IFITM3: interferon induced transmembrane protein 3 (1–8 U) | ck570309 | b8 | 0.634 | 3.021 | |
| PSMA7: proteasome (prosome, macropain) subunit, alpha type, 7 | |||||
| Cell receptor | PIGR: polymeric immunoglobulin receptor | ck570301, | a28 | 2.309 | |
| ck570303, | a39 | 2.552 | |||
| ck570313, | b13 | 1.554 | 0.330 | ||
| ck570316, | b21 | 1.869 | 0.505 | ||
| ck570318, | b24 | 1.982 | 0.394 | ||
| ck570319 | b40 | 2.942 | 0.662 | ||
| Others | C6orf62: chromosome 6 open reading frame 62 | ck570289 | a2 | 0.236 | |
| IRA1: likely ortholog of mouse IRA1 protein | ck570291 | a4 | 0.402 | ||
| MRNA: cDNA DKFZp686P07216 (from clone DKFZp686P07216) | ck570296 | a12 | 0.450 | ||
| DACH1: dachshund homolog 1 (Drosophila) | ck570298 | a14 | 0.482 | ||
| TXNIP: thioredoxin interacting protein | ck570317 | b23 | 1.959 | 0.297 | |
| LOC376745: AG1 | ck570328 | c5 | 0.336 | ||
| Transcribed sequence with strong similarity to protein prf:0512543A (H. sapiens) 0512543A oxidase II, cytochrome | ck570333 | c12 | 0.373 | ||
| C14orf112: chromosome 14 open reading frame 112 | ck570337 | c19 | 0.414 | ||
| Transcribed sequence with moderate similarity to | ck570348 | c53 | 2.152 | ||
| protein sp:P18124 (H. sapiens) RL7_HUMAN 60S | ck570354 | c71 | 2.648 | ||
| ribosomal protein L7 | ck570358 | c82 | 3.127 | ||
| F LJ32421: hypothetical protein FLJ32421 | ck570364, | c50 | 2.104 | ||
| C20orf45: chromosome 20 open reading frame 45 | ck570322, | b38 | 2.665 | 0.368 | |
| ck570327, | b44 | 2.942 | 0.590 | ||
| CDNA clone MGC:62026 IMAGE:6450688, complete cds | ck570330, | c10 | 0.368 | ||
HCRAC is a multi-gene disease similar to other malignant tumors[1]. Although the expression of some genes such as APC, DCC and p53 is related with the occurrence of HCRAC, the underlying mechanisms and the whole gene expression profile involved in the pathogenesis of HCRAC are still unclear[5-9]. In this study, according to the SSH technique based on suppression PCR[10], the cDNA subtractive library of HCRAC was constructed. The subtraction efficiency was estimated by PCR analysis. The amount of G3PDH was relatively decreased and the amount of c-myc was relatively increased. Thirty-six randomly selected clones were sequenced. Bioinformatics analyses showed human ESTs and dbEST with their accession numbers in the GenBank[11].
SSH combined with cDNA microarray or real-time-PCR technique has been used as an efficient method to identify differently expressed genes in breast cancer cells, renal cells, colon cancer cells, non-metastatic and metastatic cancer cells, lung cancer cells, bronchial epithelial cells, fibroblast growth factor 2-transformed endothelial cells[12-19]. It was reported that some differently expressed tissue-specific genes have been identified in nasopharyngeal epithelial tissue and two novel full-length genes have been isolated in human glioma specimens[20,21].
Cancer is a highly variable disease with multiple heterogeneous genetic and epigenetic changes. Functional studies are essential to understand the complex polymorphisms of cancer. Microarray is a new powerful tool for studying the molecular basis of interaction. This technique makes it possible to examine the expression of thousands of genes simultaneously[23]. To further confirm the different expression of genes identified in subtractions in the present study, four pairs of samples of patients were chosen for cDNA microarray analysis. The results of the forward and reverse hybridizations were identical.
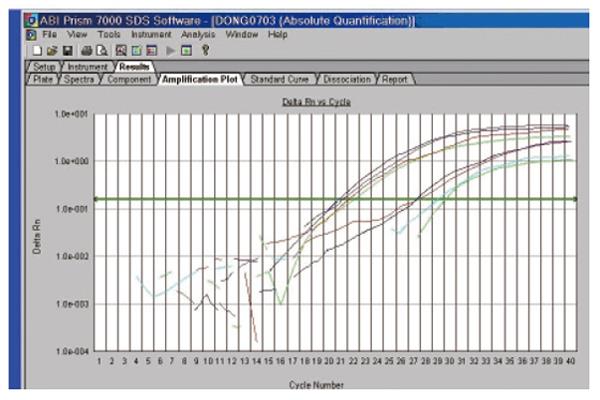
Both thermodynamic and physical parameters are known to influence hybridization intensities on DNA microarray, and they may fail in discriminating highly similar genes within a gene family during heterologous probe hybridization. Among the techniques available to validate the DNA microarray data, we chose quantitative real-time RT-PCR (qRT-PCR), since it could offer confirmatory quantitative results under stringent conditions. qRT-PCR can be used for measuring low concentrations of mRNA, and is highly accurate, reproducible, and amenable to high throughput analysis[23-31]. Three differently expressed ESTs were selected and their expression was analyzed by reverse transcriptase real-time quantitative PCR in the ten pairs of patients. The expression of the four cDNA fragments tested displayed the expected different patterns of expression (Figure 5).
Carcinogenesis is a complex process in which many molecules are changed at transcriptional and translational levels, and then manifested as structural and functional changes of cancer cells. In forward microarray hybridization, eukaryotic translation initiation factor 3 and SRY Sox are downregulated. Furthermore, heat shock proteins 10 and 60 KD, ribosomal protein S 17, ribosomal protein S 25, ribosomal protein S 28, ribosomal protein L7, tubulin alpha and beta, E-cadherin and integrin alpha 6 proteasome are transiently downregulated. The up-regulated calmodulin 1, nuclear gene encoding mitochondrial protein, protease, ATPase, Na+/K+ beta transporting polypeptide, Zinc finger protein, and putative ribosomal RNA apurinic site-specific lyase have been demonstrated in forward microarray hybridization. The results of reverse microarray hybridization coincided with it.
Translation initiation in eukaryotes is a rate-limiting step in protein synthesis. It is a complex process involving many eukaryotic initiation factors (eIFs). Altering the expression level or the function of eIFs may influence the synthesis of some proteins and consequently cause abnormal cell growth and malignant transformation[32]. Ribosomes that catalyze synthesis of proteins consist of a small 40S subunit and a large 60S subunit. These subunits are composed of four RNA species and approximately 80 structurally distinct proteins[33]. The down-regulated tubulin has been observed. This experiment renders further support to the hypothesis that activation of both the transcriptional and posttranscriptional machinery may constitute an integral part of the mechanism underlying carcinogenesis.
Down-regulated E-cadherin and integrin alpha 6 can decrease cancer cell adhesion and increase its invasion. Up-regulated nuclear genes encoding mitochondrial protein, ATPase, Na+/K+ beta transporting polypeptide reflect that cancer cells need enough energy to grow.
We observed that Sox9 was strongly down-regulated (0.426-fold). Sox, a high-mobility-group domain containing transcription factor, with a DNA-binding domain similar to that of the mammalian testis-determining factor SRY, is a key transcription factor that is essential for chondrocyte differentiation and chondrocyte-specific gene expressions[34,35]. Sox9 has been demonstrated to be a “master regulator” gene that controls distinct pathways of mesenchymal differentiation. Sox9 is a target of cAMP signaling and phosphorylation of Sox9 by protein kinase A (PKA) enhances its transcriptional and DNA-binding activity[36-38]. Sox9 harbors a number of highly conserved regions, including two domains required for maximal trans-activation. The heat shock protein HSP70 recognizes a specific region of Sox9 with unknown function which may facilitate the assembly of multi-protein complexes at promoter enhancer regions. The Sox9 HMG domains carry two nuclear localization signals (NLSs). The N-terminal NLS binds to calmodulin while the C-terminal NLS binds to importin beta[39].
We also found that heat shock protein 10 KD (chaperonin 10) was downregulated (0.562-fold). Heat shock protein 10 KD can reduce myocyte death by its mitochondrial function or by interacting with cytoplasmic signaling pathways. The Ras GTPase signaling pathway indicates that inhibition of Ras is required for the protection by HSP10. In abnormal situations, oncogenic activation of Ras signal transduction pathways leads to continuous upregulation of key elements of translational machinery. The Ras signal transduction pathways play a critical role in regulating mRNA translation and cellular transformation. On the other hand, tumor suppressor genes downregulate ribosomal and tRNA synthesis, and their inactivation results in uncontrolled production of these translational components. Heat shock proteins (chaperonins) are also a subgroup of oligomeric molecular chaperones. Chaperonins 10 and 60 can be found on the surface of various prokaryotic and eukaryotic cells, and release from cells. Secreted chaperonins can interact with a variety of cell types, including leukocytes, vascular endothelial and epithelial cells, and activate key cellular activities such as the synthesis of cytokines and adhesion proteins[40-42]
We further observed that some receptor subunits (calmodulin) related to ion channels (potassium channel protein) were strongly upregulated (3.265-fold). Calmodulin (phosphorylase kinase delta) is a major cytoplasmic calcium receptor that performs multiple functions in cells including cytokinesis[43]. Calmodulin is a small protein involved in calcium signaling and is able to bind to many different targets. The targets of calmodulin are a number of kinases, including myosin light chain kinase (MLCK), calmodulin-dependent kinase and phosphorylase kinase. The phosphorylase kinase holoenzyme (PhK) including alpha-beta, gamma and delta alters the interaction between its regulatory alpha and catalytic gamma subunits. The gamma subunit is also known to interact with the delta subunit, an endogenous molecule of calmodulin that mediates the activation of PhK by Ca (2+) ions[44].
In conclusion, the down-regulated transcription factor Sox9 can influence cell differentiation and cell-specific gene expression. Down-regulation of heat shock protein 10 KD loses its inhibition of Ras, and attenuates the Ras GTPase signaling pathway, increases cell proliferation and inhibits cell apoptosis, which is a hallmark of aggressive malignant colorectal adenocarcinoma. Down-regulated Sox9 also decreases its binding to calmodulin, accumulates calmodulin as receptor-activated kinase and phosphorylase kinase due to the activation of PhK by Ca (2+) ions. Upregulated nuclear DNA-encoded mitochondria supply increases cancer cell proliferation energy.
| 1. | Chan WM, Pang CP, Lam DS. Genetics of colorectal cancer. N Engl J Med. 2003;348:2361-2362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (1)] |
| 2. | Cho KR, Vogelstein B. Genetic alterations in the adenoma--carcinoma sequence. Cancer. 1992;70:1727-1731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 3. | Benhattar J, Losi L, Chaubert P, Givel JC, Costa J. Prognostic significance of K-ras mutations in colorectal carcinoma. Gastroenterology. 1993;104:1044-1048. [PubMed] |
| 4. | Smith DR, Myint T, Goh HS. Over-expression of the c-myc proto-oncogene in colorectal carcinoma. Br J Cancer. 1993;68:407-413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 63] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Tokuda T, Tamaoka A, Matsuno S, Sakurai S, Shimada H, Morita H, Ikeda S. Plasma levels of amyloid beta proteins did not differ between subjects taking statins and those not taking statins. Ann Neurol. 2001;49:546-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 6. | Hermeking H, Eick D. Mediation of c-Myc-induced apoptosis by p53. Science. 1994;265:2091-2093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 546] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 7. | Bonneton C, Larue L, Thiery JP. [Current data on the role of APC protein in the origin of colorectal cancer]. Bull Cancer. 1997;84:1053-1060. [PubMed] |
| 8. | Peltomäki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet. 2001;10:735-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 318] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 9. | Marcus VA, Madlensky L, Gryfe R, Kim H, So K, Millar A, Temple LK, Hsieh E, Hiruki T, Narod S. Immunohistochemistry for hMLH1 and hMSH2: a practical test for DNA mismatch repair-deficient tumors. Am J Surg Pathol. 1999;23:1248-1255. [PubMed] |
| 10. | Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996;93:6025-6030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2262] [Cited by in RCA: 2004] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 11. | Yao Chen, Yizheng Zhang. The construction of cDNA suppression subtractive library of human rectum adenocarcinomas. J U S Lymph Oncogy. 2002;1:9-14. |
| 12. | Yang GP, Ross DT, Kuang WW, Brown PO, Weigel RJ. Combining SSH and cDNA microarrays for rapid identification of differentially expressed genes. Nucleic Acids Res. 1999;27:1517-1523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 170] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 13. | Stassar MJ, Devitt G, Brosius M, Rinnab L, Prang J, Schradin T, Simon J, Petersen S, Kopp-Schneider A, Zöller M. Identification of human renal cell carcinoma associated genes by suppression subtractive hybridization. Br J Cancer. 2001;85:1372-1382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 117] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 14. | Swearingen ML, Sun D, Bourner M, Weinstein EJ. Detection of differentially expressed HES-6 gene in metastatic colon carcinoma by combination of suppression subtractive hybridization and cDNA library array. Cancer Lett. 2003;198:229-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 15. | Goldberg SF, Miele ME, Hatta N, Takata M, Paquette-Straub C, Freedman LP, Welch DR. Melanoma metastasis suppression by chromosome 6: evidence for a pathway regulated by CRSP3 and TXNIP. Cancer Res. 2003;63:432-440. [PubMed] |
| 16. | Sers C, Tchernitsa OI, Zuber J, Diatchenko L, Zhumabayeva B, Desai S, Htun S, Hyder K, Wiechen K, Agoulnik A. Gene expression profiling in RAS oncogene-transformed cell lines and in solid tumors using subtractive suppression hybridization and cDNA arrays. Adv Enzyme Regul. 2002;42:63-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Bangur CS, Switzer A, Fan L, Marton MJ, Meyer MR, Wang T. Identification of genes over-expressed in small cell lung carcinoma using suppression subtractive hybridization and cDNA microarray expression analysis. Oncogene. 2002;21:3814-3825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Nishikawa J, Kiss C, Imai S, Takada K, Okita K, Klein G, Szekely L. Upregulation of the truncated basic hair keratin 1(hHb1-DeltaN) in carcinoma cells by Epstein-Barr virus (EBV). Int J Cancer. 2003;107:597-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 19. | An Q, Pacyna-Gengelbach M, Schlüns K, Deutschmann N, Guo S, Gao Y, Zhang J, Cheng S, Petersen I. Identification of differentially expressed genes in immortalized human bronchial epithelial cell line as a model for in vitro study of lung carcinogenesis. Int J Cancer. 2003;103:194-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Qi ZY, Li Y, Ying K, Wu CQ, Tang R, Zhou ZX, Chen ZP, Hui GZ, Xie Y. Isolation of novel differentially expressed genes related to human glioma using cDNA microarray and characterizations of two novel full-length genes. J Neurooncol. 2002;56:197-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 21. | Zhang B, Nie X, Xiao B, Xiang J, Shen S, Gong J, Zhou M, Zhu S, Zhou J, Qian J. Identification of tissue-specific genes in nasopharyngeal epithelial tissue and differentially expressed genes in nasopharyngeal carcinoma by suppression subtractive hybridization and cDNA microarray. Genes Chromosomes Cancer. 2003;38:80-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 57] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 22. | Russo G, Zegar C, Giordano A. Advantages and limitations of microarray technology in human cancer. Oncogene. 2003;22:6497-6507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 163] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 23. | Qiu J, Gunaratne P, Peterson LE, Khurana D, Walsham N, Loulseged H, Karni RJ, Roussel E, Gibbs RA, Margolin JF. Novel potential ALL low-risk markers revealed by gene expression profiling with new high-throughput SSH-CCS-PCR. Leukemia. 2003;17:1891-1900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 24. | Ahmed FE. Molecular techniques for studying gene expression in carcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2002;20:77-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 25. | Dooley TP, Reddy SP, Wilborn TW, Davis RL. Biomarkers of human cutaneous squamous cell carcinoma from tissues and cell lines identified by DNA microarrays and qRT-PCR. Biochem Biophys Res Commun. 2003;306:1026-1036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 61] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 26. | Chuang YY, Chen Y, Gadisetti VR, Cook JA, Coffin D, Tsai MH, DeGraff W, Yan H, Zhao S, Russo A. Gene expression after treatment with hydrogen peroxide, menadione, or t-butyl hydroperoxide in breast cancer cells. Cancer Res. 2002;62:6246-6254. [PubMed] |
| 27. | Pagliarulo V, George B, Beil SJ, Groshen S, Laird PW, Cai J, Willey J, Cote RJ, Datar RH. Sensitivity and reproducibility of standardized-competitive RT-PCR for transcript quantification and its comparison with real time RT-PCR. Mol Cancer. 2004;3:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 28. | Pinhasov A, Mei J, Amaratunga D, Amato FA, Lu H, Kauffman J, Xin H, Brenneman DE, Johnson DL, Andrade-Gordon P. Gene expression analysis for high throughput screening applications. Comb Chem High Throughput Screen. 2004;7:133-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 29. | Luthra R, Sanchez-Vega B, Medeiros LJ. TaqMan RT-PCR assay coupled with capillary electrophoresis for quantification and identification of bcr-abl transcript type. Mod Pathol. 2004;17:96-103. [PubMed] |
| 30. | Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24144] [Cited by in RCA: 26611] [Article Influence: 1064.4] [Reference Citation Analysis (0)] |
| 31. | Li Q, Luan G, Guo Q, Liang J. A new class of homogeneous nucleic acid probes based on specific displacement hybridization. Nucleic Acids Res. 2002;30:E5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 210] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 32. | Dong Z, Liu LH, Han B, Pincheira R, Zhang JT. Role of eIF3 p170 in controlling synthesis of ribonucleotide reductase M2 and cell growth. Oncogene. 2004;23:3790-3801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 86] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 33. | Kenmochi N, Kawaguchi T, Rozen S, Davis E, Goodman N, Hudson TJ, Tanaka T, Page DC. A map of 75 human ribosomal protein genes. Genome Res. 1998;8:509-523. [PubMed] |
| 34. | Lefebvre V, de Crombrugghe B. Toward understanding SOX9 function in chondrocyte differentiation. Matrix Biol. 1998;16:529-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 206] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 35. | Tsuda M, Takahashi S, Takahashi Y, Asahara H. Transcriptional co-activators CREB-binding protein and p300 regulate chondrocyte-specific gene expression via association with Sox9. J Biol Chem. 2003;278:27224-27229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 166] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 36. | Wehrli BM, Huang W, De Crombrugghe B, Ayala AG, Czerniak B. Sox9, a master regulator of chondrogenesis, distinguishes mesenchymal chondrosarcoma from other small blue round cell tumors. Hum Pathol. 2003;34:263-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 131] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 37. | Huang W, Zhou X, Lefebvre V, de Crombrugghe B. Phosphorylation of SOX9 by cyclic AMP-dependent protein kinase A enhances SOX9's ability to transactivate a Col2a1 chondrocyte-specific enhancer. Mol Cell Biol. 2000;20:4149-4158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 217] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 38. | Afonja O, Raaka BM, Huang A, Das S, Zhao X, Helmer E, Juste D, Samuels HH. RAR agonists stimulate SOX9 gene expression in breast cancer cell lines: evidence for a role in retinoid-mediated growth inhibition. Oncogene. 2002;21:7850-7860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 65] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 39. | Harley VR. The molecular action of testis-determining factors SRY and SOX9. Novartis Found Symp. 2002;244:57-66; discussion 66-7, 79-85, 253-7. [PubMed] |
| 40. | Lin KM, Hollander JM, Kao VY, Lin B, Macpherson L, Dillmann WH. Myocyte protection by 10 kD heat shock protein (Hsp10) involves the mobile loop and attenuation of the Ras GTP-ase pathway. FASEB J. 2004;18:1004-1006. [PubMed] |
| 41. | Ranford JC, Coates AR, Henderson B. Chaperonins are cell-signalling proteins: the unfolding biology of molecular chaperones. Expert Rev Mol Med. 2000;2:1-17. [PubMed] |
| 42. | Rosenwald IB. The role of translation in neoplastic transformation from a pathologist's point of view. Oncogene. 2004;23:3230-3247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 95] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 43. | Yu YY, Chen Y, Dai G, Chen J, Sun XM, Wen CJ, Zhao DH, Chang DC, Li CJ. The association of calmodulin with central spindle regulates the initiation of cytokinesis in HeLa cells. Int J Biochem Cell Biol. 2004;36:1562-1572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 44. | Yang C, Jas GS, Kuczera K. Structure, dynamics and interaction with kinase targets: computer simulations of calmodulin. Biochim Biophys Acta. 2004;1697:289-300. [PubMed] |
S- Editor Guo SY L- Editor Wang XL E- Editor Cao L