Published online Dec 7, 2006. doi: 10.3748/wjg.v12.i45.7278
Revised: April 28, 2006
Accepted: October 10, 2006
Published online: December 7, 2006
AIM: To determine the presence of Helicobacter species DNA in the liver of chronic hepatitis C (CHC) patients with and without cirrhosis as compared to controls, and to identify the bacterial species involved.
METHODS: Seventy-nine consecutive patients (HBV and HIV negative) with a liver sample obtained after liver biopsy or hepatic resection were studied: 41 with CHC without cirrhosis, 12 with CHC and cirrhosis, and 26 controls (HCV negative). Polymerase chain reactions (PCRs) targeting Helicobacter 16S rDNA and species-specific were performed on DNA extracted from the liver. A gastric infection with H pylori was determined by serology and confirmed by 13C-urea breath test.
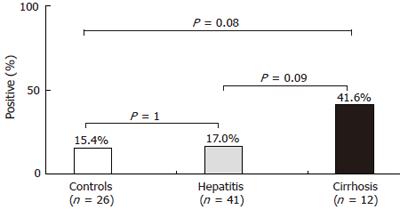
RESULTS: Overall, Helicobacter 16S rDNA was found in 16 patients (20.2%). Although positive cases tended to be higher in CHC patients with cirrhosis (41.6%) than in those without cirrhosis (17.0%) or in controls (15.4%), the difference was not statistically significant (P = 0.08). H pylori-like DNA was identified in 12 cases and H. pullorum DNA in 2, while 2 cases remained unidenti-fied. Gastric infection with H pylori was found in only 2 of these patients.
CONCLUSION: Our results do not confirm the associ-ation of Helicobacter species DNA in the liver of CHC patients with advanced liver disease. The lack of correlation between positive H pylori serology and the presence of H pylori-like DNA in the liver may indicate the presence of a variant of this species.
-
Citation: Castéra L, Pedeboscq A, Rocha M, Bail BL, Asencio C, Lédinghen V, Bernard PH, Laurent C, Lafon ME, Capdepont M, Couzigou P, Bioulac-Sage P, Balabaud C, Mégraud F, Ménard A. Relationship between the severity of hepatitis C virus-related liver disease and the presence of
Helicobacter species in the liver: A prospective study. World J Gastroenterol 2006; 12(45): 7278-7284 - URL: https://www.wjgnet.com/1007-9327/full/v12/i45/7278.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i45.7278
Chronic hepatitis C virus (HCV) infection is a major public health problem with over 170 million people infected worldwide. It is the leading cause of chronic liver disease and the main indication for liver transplantation in the Western world[1]. Approximately 80% of patients with acute infection develop chronic hepatitis. Chronic hepatitis C is associated with a wide spectrum of liver histological lesions ranging from mild chronic hepatitis to cirrhosis and hepatocellular carcinoma (HCC)[2]. The course of HCV-related hepatic disease varies markedly from one patient to another. Several factors including age at exposure, duration of infection, alcohol intake, male gender and more recently steatosis have been shown to be associated with fibrosis progression[3-6]. However, even in the absence of these factors, disease progression may be observed in some patients, suggesting the role of other factors which remain to be identified. Host genetic factors or environmental factors, such as a bacterial coinfection, could be involved.
Several Helicobacter species colonize the liver of animals and induce hepatitis[7]. In the past few years, the emergence of new Helicobacter species associated with the pathogenesis of human enterohepatic diseases has been observed[8-10]. H pylori and Helicobacter pullorum (H. pullorum) DNA have been detected in the liver tissue of patients with chronic hepatitis C and HCC, suggesting that these bacteria could be implicated in the progression of chronic hepatitis C to cirrhosis and HCC[11]. In addition, we have shown recently in a cross-sectional retrospective study, a significant association between the presence of Helicobacter species DNA in the liver and HCV-related cirrhosis with and without HCC[12]. Given the limitation of retrospective studies, the aim of this new study was to prospectively determine the prevalence of Helicobacter infection of the liver in HCV-infected patients with and without cirrhosis as compared to controls.
All patients with chronic hepatitis C undergoing a percutaneous liver biopsy at the Hepatology Clinics of Bordeaux University between March 2002 and December 2003 were eligible. Chronic hepatitis C was defined by detection of HCV antibodies using a third generation test (Ortho HCV 3.0 ELISA Monolisa anti-HCV, BioRad, Marne la Coquette, France), detectable HCV RNA (Cobas amplicor HCV 2.0, Roche Diagnostics, Neuilly sur Seine, France) in serum, and elevated alanine aminotransferase (ALT) levels for more than 6 mo. Exclusion criteria were age under 18 years, co-infection with hepatitis B virus (HBV) or human immunodeficiency virus (HIV).
The following data were collected at the time of enrolment of patients with chronic HCV infection: age, gender, route of HCV transmission, duration of HCV infection, alcohol intake, aspartate aminotransferase (AST), ALT, gamma-glutamyl transpeptidase (GGT), HCV genotype and viral load.
All patients seronegative for HCV and undergoing hepatic resection for benign tumors or metastatic tumors during the study period, were eligible and served as controls. All patients gave their informed consent and the study protocol conformed to the 1975 declaration of Helsinki and was approved by the Ethics Committee of our institution.
Fresh liver tissues obtained either from needle biopsies or from surgical specimens during usual diagnostic or therapeutic procedures, were immediately cut into three parts with three different conditionning protocols: formalin fixation for routine histology (at least 1 cm for needle biospies), freezing in liquid nitrogen-cooled isopentane followed by storage at -80°C before molecular biology, and immersion in a specific culture medium for culture characterization of the bacterium.
For all patients, conventional liver histology was performed on formalin-fixed liver tissues. Sections were stained with hematein-eosin-safran, Masson’s trichromic stain and reticulin stain. In patients with chronic hepatitis C, liver fibrosis and necroinflammatory activity were evaluated semi-quantitatively according to the METAVIR scoring system[13]. Fibrosis was staged on a 0-4 scale as follows: F0 = no fibrosis; F1 = portal fibrosis without septa; F2 = portal fibrosis and few septa; F3 = numerous septa without cirrhosis; F4 = cirrhosis. Activity was graded as follows: A0 = none; A1 = mild; A2 = moderate; A3 = severe. The diagnosis of hepatocellular carcinoma was based on usual criteria[14].
Serum samples from all patients were tested for anti H pylori IgG antibodies, using the commercially available kit Pyloriset EIA-GIII(Orion Diagnostica, Espoo, Finland) and the Western blot assay HELICO BLOT 2.1 (Genelabs Diagnostics®, Singapore). When antibodies were present, gastric infection with H pylori was searched by 13C-urea breath test (UBT) or by culture on gastric biopsy specimens obtained during upper gastrointestinal endoscopy when indicated for diagnosis of esophageal varices in patients with cirrhosis. UBT was performed after overnight fasting, 75 mg of urea was ingested after citric acid. Air samples were obtained before and 30 min after urea ingestion and analyzed by isotope ratio mass spectrometry. Serology and UBT have 95% sensitivity and specificity[15].
Standard Helicobacter culture methods were used including Wilkins Chalgren agar with antibiotics[16] and chocolate agar without antibiotics. The plates with fresh liver tissues were incubated at 37°C for 4 d, one set in a microaerobic atmosphere and another in an anaerobic atmosphere.
To maximize the culture of Helicobacter from the biopsies, a flask of confluent murine hepatic (CCl 9.1) cells was systematically inoculated with liver samples and observed during a week for a color change or opacification of the medium, or for a cytopathic effect. The medium was also observed microscopically before discarded.
DNA from frozen liver material (20 to 25 mg/specimen) was extracted by using the QIAamp kit (Qiagen Inc., Chatsworth, CA) as previously described[12].
Standard PCR amplifications were carried out as previously reported and PCR products were analyzed on a 1%-4% agarose gel, depending on the amplicon size, and stained with ethidium bromide[12].
Two real time PCRs were performed using the TaqMan® or SYBR® Green chemistries. The TaqMan real time PCR amplification and hybridization reactions were carried out in a final volume of 10 μL containing 5 μL of TaqMan® Universal PCR Master Mix, 0.3 μmol/L of each primer, 0.2 μmol/L of the labeled probe Helico-spp-16S and 1 μL of purified DNA in a ABI PRISM® 7000 thermocycler (Applied Biosystems, Foster City, CA). DNA was amplified using the following cycling parameters: heating at 95°C for 10 min followed by 40 cycles of a two-stage temperature profile of 95°C for 15 s and 60°C for 1 min. The SYBR® Green real time PCR amplification and melting curve analysis were carried out in a final volume of 10 μL containing 5 μL of SYBR® Green PCR Master Mix, 0.3 μmol/L of each primer and 1 μL of purified DNA in a ABI PRISM® 7000 thermocycler (Applied Biosystems). DNA was amplified using the following cycling parameters: heating at 50°C for 2 min, then at 95°C for 10 min followed by 40 cycles of a two-stage temperature profile of 95°C for 15 s and 60°C for 1 min. Then a dissociation of the amplicon from 60°C to 95°C was followed during 20 min.
A PCR targeting a 130-bp sequence of the malate deshydrogenase (mdh) gene of E. coli was applied to the liver tissue as previously described[12].
Helicobacter genus-specific primer pairs C97/C98[17] and HS1/HS2[8,18] were used to generate 16S rDNA amplicons of approximately 400 bp. Moreover, a new PCR targeting the Helicobacter genus 16S rRNA gene was developed using the TaqMan chemistry. Sequences of different Helicobacter species such as H pylori, Helicobacter heilmannii, Helicobacter fennelliae, Flexispira rappini, Helicobacter bilis, Helicobacter cinaedi, Helicobacter winghamensis, Helicobacter pullorum, and Helicobacter bizzozeronii were aligned. Closely related 16S rRNA genes from various bacteria, such as Campylobacter coli, Campylobacter jejuni, Campylobacter fetus, Wolinella succinogenes, were also included and a search was carried out to identify conserved regions specific to the Helicobacter genus. As a result, a set of primers (AS2-Helico-TQM: 5’- CCGTGTCTCAGTTCCAGTGTGT-3’ and S1-Helico-TQM: 5’- GATCAGCCTATGTCCTATCAGCTTGT-3’) were designed to amplify a 106-bp sequence in Helicobacter species. After the specificity of the PCR was ensured, a probe was also designed (Helico-spp-16S FAM-TCACCCTCTCAGGCCGGATACCC-TAMRA). TaqMan probes were synthesized with the FAM reporter dye covalently linked to the 5’P ends and the TAMRA quencher dye at the 3’P ends which were phosphorylated to prevent probe extension. Primers and probes used for the TaqMan assays synthesized by Proligo (Paris, France) were designed using a Primer Express software package PE-ABI (Applied Biosystems).
Samples generating a positive result with the Helicobacter genus-specific PCR were subsequently analyzed with seven different sets of primers for the detection of four species previously found in human liver, i.e. H. sbilis, H. spullorum, H pylori and F. rappini[12]. For H. sbilis and H. spullorum primers amplifying a 151-bp product and a 140-bp product on the cdtB gene were used, respectively[12]. For F. rappini, primers amplifying a 101-bp product on the ureB gene were used[12]. For H pylori, four real time PCRs were performed for detecting a 267-bp product on the 23S rRNA gene[19], a 146-bp product on the glmM (ureC) gene[20], a 231-bp product on the ureA gene[21], and a 303-bp product on the 26 kDa specific antigen gene[22].
Given the possibility of the presence of different strains in the same sample, PCR products were cloned prior to sequencing. Helicobacter species 16S rDNA samples were amplified, cloned and sequenced as previously reported[12]. Helicobacter genus-specific and species-specific amplified primer-less sequences were compared to the GenBank database with the Blast program at the National Center for Biotechnology Information Computer server[23].
Descriptive statistics are provided as means ± SD. One way analysis of variance (ANOVA) or the Mann-Whitney test was used when necessary for statistical comparison of quantitative data, and the chi-square test or Fisher’s exact test for qualitative data. P < 0.05 was considered statistically significant.
A total of 79 patients with a liver sample obtained after percutaneous liver biopsy or hepatic resection were included. The median length of the needle biopsy specimens was 1.4 cm (mean 1.35 cm). The characteristics of the patients are shown in Table 1.
| Characteristics | Group 1(Hepatitis) | Group 2(Cirrhosis) | Group 3(Controls) |
| Patients (n) | 41 | 12 | 26 |
| Age (yr) | 50 ± 13 | 60 ± 11a | 49 ± 12 |
| Gender (M/F) | 14/27 | 5/7 | 13/13 |
| Route of transmission, n (%) | |||
| Transfusion | 10 (24%) | 5 (42%) | - |
| IVDU | 11 (27%) | 3 (25%) | - |
| Other or unknow | 20 (49%) | 4 (33%) | - |
| Duration of infection (yr) | 19 ± 9 | 23 ± 5 | - |
| Alcohol intake (g/d) | 12 ± 23 | 12 ± 14 | - |
| AST (xULN) | 1.5 ± 1.1 | 3.7 ± 2.4b | - |
| ALT (xULN) | 1.7 ± 1.3 | 3.7 ± 2.8b | - |
| GGT (xULN) | 1.4 ± 0.7 | 2.0 ± 1.9 | - |
| HCV genotype, n (%) | - | ||
| 1 | 24 (58%) | 8 (67%) | - |
| 2/3 | 11 (27%) | 4 (33%) | - |
| Other | 6 (15%) | 0 (0%) | - |
| HCV viral load (IU/mL) | 827 889 ± 123 741 | 923 026 ± 181 866 | - |
| Histological lesions (Metavir) | |||
| Activity, n (%) | |||
| A0-1 | 16 (39%) | 0 (0%) | - |
| A2-3 | 25 (61%) | 12 (100%)a | - |
| Fibrosis, n (%) | |||
| F0-1 | 16 (39%) | - | - |
| F2 | 23 (56%) | - | - |
| F3 | 2 (5%) | - | - |
| F4 | - | 12 (100%) | - |
Fifty-three patients who had chronic hepatitis C were divided into two groups: group 1 consisting of patients without cirrhosis (n = 41), group 2 consisting of patients with cirrhosis (n = 12). One patient in group 2 also had HCC. As expected, patients with cirrhosis (group 2) were significantly older than those without cirrhosis (group 1) (60 ± 11 years vs 50 ± 13 years, respectively; P < 0.03) and had significantly higher AST levels (3.7 ± 2.4 upper limit of normal value vs 1.5 ± 1.1 upper limit of normal value, respectively; P = 0.001) and ALT levels (3.7 ± 2.8 ULN vs 1.7 ± 1.3 ULN, respectively; P < 0.003). Conversely, they did not differ in gender distribution, route of HCV transmission, duration of infection, alcohol intake, GGT levels, HCV genotype and viral load.
Group 3 consisted of HCV negative control patients (n = 26) in whom liver tissues were taken from the non-tumoral part of hepatectomy specimens after resection for hepatic benign tumors (n = 11; i.e. 4 focal nodular hyperplasias, 2 hydatic cysts, 1 cavernous hemangioma, 1 liver cell adenoma, 1 biliary cyst, 1 cystadenoma, 1 abcess) or metastatic tumors (n = 15; i.e. all 15 of colorectal origin).
Despite our efforts, it was impossible to grow any Helicobacter strain from the liver, either on the plate media or in the tissue culture flasks.
Overall, Helicobacter genus DNA was detected in the liver of 16 patients (20.2%). These positive cases were found in 7/41 (17.0%) patients of group 1, 5/12 (41.6%) patients of group 2, and 4/26 (15.4%) patients of group 3 (Figure 1). Although positive cases tended to be more frequent in patients with cirrhosis than in those without cirrhosis or in controls, the difference did not reach statistical significance (P = 0.08). Among the 79 liver specimens studied, none was positive by PCR for E. coli, a frequent colonizer of the gut.
A number of factors were tested for their association with the presence of Helicobacter DNA in the liver of 53 patients with chronic hepatitis C (with or without cirrhosis). None of these factors was associated with the presence of Helicobacter (Table 2). Positive patients were significantly younger than negative patients (46 ± 13 years vs 54 ± 13 years, respectively; P = 0.05).
| Factor | Helicobacter DNA | P | |
| Present | Absent | ||
| Patients (n) | 12 | 41 | |
| Age (yr) | 46 ± 13 | 54 ± 13 | 0.05 |
| Gender (M/F) | 5/7 | 12/29 | NS |
| Route of transmission | |||
| Transfusion | 3 (25%) | 12 (29%) | |
| IVDU | 3 (25%) | 11 (27%) | NS |
| Other or unknown | 6 (50%) | 18 (44%) | |
| Duration of infection (yr) (n = 36) | 18 ± 7 | 21 ± 8 | NS |
| Alcohol intake (g/d) | 5 ± 8 | 14 ± 24 | NS |
| AST (xULN) | 2.2 ± 2.1 | 1.9 ± 1.5 | NS |
| ALT (xULN) | 2.7 ± 2.9 | 2.0 ± 1.5 | NS |
| GGT (xULN) | 1.3 ± 0.5 | 1.6 ± 1.2 | NS |
| HCV genotype | |||
| 1 | 9 (75%) | 23 (56%) | |
| 2/3 | 2 (17%) | 13 (24%) | NS |
| Other | 1 (8%) | 5 (20%) | |
| HCV viral load (IU/mL) | 663 824 ± 569 933 | 972 157 ± 1 388 853 | NS |
| Histological lesions (Metavir score) | |||
| Activity | |||
| A0-1 | 4 (33%) | 12 (29%) | |
| A2-3 | 8 (67%) | 29 (71%) | NS |
| Fibrosis | |||
| F0-2 | 7 (58%) | 32 (78%) | |
| F3-4 | 5 (42%) | 9 (22%) | NS |
Overall, anti H pylori IgG antibodies were detected in 20/76 (26.3%) cases. These positive cases were found in 12/41 (29.2%) patients of group 1, 3/11 (27.0%) patients of group 2 and 5/24 (20.8%) patients of group 3. As shown in Table 3, among the 16 patients positive for Helicobacter DNA in the liver, anti H pylori IgG antibodies were detected in 2/7 (28.5%) patients of group 1, 1/4 (25.0%) patients of group 2 and 1/4 (25.0%) patients of group 3. Serology was not performed in one positive patient. All the results obtained by ELISA were also confirmed using an immunoblot specific for H pylori. Gastric infection was confirmed by UBT in the 2 positive patients of group 1 but not in the positive patient of group 2. UBT was not performed in one positive control patient.
| Groups | Patient No. | H pylori infection | H pylori PCR | H. pullorum PCR | Conclusion | ||||
| Serology | UBT | 23S rDNA | ureA | 26 kDa | glmM | cdtB | |||
| Group 1 | 1 | - | - | - | - | - | - | - | Not identified |
| (Hepatitis) | 2 | - | - | - | - | + | - | + | “H. pullorum”-like1 |
| 3 | - | - | + | + | + | - | - | “H pylori”-like1 | |
| 4 | + | + | + | + | - | - | - | “H pylori”-like | |
| 5 | + | + | + | + | + | - | - | “H pylori”-like1 | |
| 6 | - | - | - | - | - | - | - | Not identified | |
| 7 | - | - | + | - | - | - | - | “H pylori”-like1 | |
| Group 2 | 8 | - | - | + | + | + | - | - | “H pylori”-like1 |
| (Cirrhosis) | 9 | - | - | + | + | + | + | - | “H pylori”-like1 |
| 10 | ND | - | + | + | + | + | - | “H pylori”-like | |
| 11 | - | - | - | - | + | - | - | “H pylori”-like | |
| 12 | + | - | - | - | + | - | + | “H. pullorum”-like | |
| Group 3 | 13 | - | - | - | - | + | - | - | “H pylori”-like |
| (Controls) | 14 | - | - | - | - | + | - | - | “H pylori”-like1 |
| 15 | + | ND | - | + | + | - | - | “H pylori”-like | |
| 16 | - | - | + | + | + | - | - | “H pylori”-like | |
Material from 16 Helicobacter genus positive patients was tested with species-specific primers (Table 3). Neither H. bilis nor H. rappini was found (data not shown). H pylori-like organisms were identified in 12 (75%) cases and H. spullorum-like organisms were identified in 2 cases (12.5%), 2 samples remained unidentified. H pylori-like organisms were present in 4/7 (57%) patients of group 1, 4/5 (80.0%) patients of group 2, and 4/4 (100%) patients of group 3. These results were confirmed by sequencing the 16S rRNA gene in the 7 cases where it was carried out. Among the positive samples by sequencing or by H pylori specific PCR, none of them reacted with the highly conserved primer designed to amplify the specific vacA gene of H pylori and variable results were obtained with other H pylori specific targets, such as ureA, glmM. Among the 7 samples for which the near complete 16S rRNA gene was sequenced, the 2 nucleotide polymorphisms in position 92 and 130 which were found to be associated with Helicobacter 16S rDNA sequences from the liver described by Verhoef et al[29] were detected in 5 instances: 3 sequences harboured the 2 polymorphisms simultaneously, 2 sequences harboured one of these 2 polymorphisms and 2 sequences did not present these polymorphisms.
In a previous retrospective cross-sectional study[12], we showed that DNA from H pylori- and H. spullorum-like organisms was present in the liver of patients with hepatitis C cirrhosis with or without HCC, suggesting that Helicobacter species could be a co-risk factor for progression of HCV chronic liver diseases. However, a limitation of this preliminary study was its retrospective nature which did not allow us to determine whether H pylori was present in the stomach of these patients and to gather all of the clinical and biological information on HCV genotype and duration of infection, etc, which is needed for a more accurate analysis.
In this prospective study, we were able to better characterize the presence of Helicobacter species in the liver of a group of consecutive patients with HCV infection with or without cirrhosis, using both the tools which were developed in the retrospective study as well as new tools.
The results of the present study only partially confirm those obtained previously. Although the prevalence of Helicobacter DNA tended to be higher in the liver samples from patients with hepatitis C cirrhosis than in those from HCV-infected patients without cirrhosis or from controls, the difference did not reach statistical significance (P = 0.09 and P = 0.08, respectively). However, we cannot exclude the fact that given the limited number of patients with HCV- induced cirrhosis, this result may be due to a lack of statistical power. The negative results obtained with regard to the detection of E. coli DNA in liver material confirm that H pylori DNA detected is not the result of a non specific transport or failure in elimination by a non functional liver. Indeed, since E. coli is constantly present in the human intestine, if a non specific bacterial translocation occurred, we could find E. coli DNA in the liver.
This study did not enable us to solve the issue of identifying specific Helicobacter species involved since no positive culture occurred on the media usually used for Helicobacter culture. Indeed, in another study bacteria closely related to H pylori morphologically were visualized in the liver of 6 out of 20 patients with HCC[24]. Furthermore, Oliveira et al only found H pylori in two studies on ulcerative colitis[25] and Crohn’s disease patients[26]. For the latter, H pylori was more frequent than in the intestinal mucosa of the control group. These two studies were however carried out in a country of high H pylori prevalence, Brazil.
In the current study, we also had the possibility to perform H pylori serology on most patients. Interestingly, among the 15 patients (out of 16) positive for Helicobacter DNA in the liver, only 4 (26.6%) had H pylori antibodies. Given this surprising result, immunoblot was performed and the results were confirmed.
Unfortunately, the protocol did not include a UBT when Helicobacter DNA was present in the liver, but only when H pylori serology was positive. Indeed, 2 out of 3 positive serology cases were confirmed by UBT. As the sensitivity of our serological kit is 95%[15,27], it is surprising that such a high number of patients (11/15) did not produce H pylori antibodies, which casts a doubt on the identification of the species present. Two potential explanations can be given: (1) H pylori is truly present in the liver, but not in the stomach, and at an insufficient load to stimulate an immune response; and (2) another Helicobacter species which is closely related to H pylori but does not cross-react with H pylori antigens, is present in the liver. These patients did not receive H pylori eradication therapy, which could explain the eradication of the bacteria. The possibility of immunodeficiency in HCV positive patients, especially with cirrhosis can also be considered as an explanation of the lack of serological response but we have not found arguments for this hypothesis.
The 1370 bp sequence of the 16S rRNA gene of six patients with H pylori DNA shows 99% homology with H pylori, but it is known that 16S rDNA sequences do not have a good discriminatory power in the Helicobacter genus. A recent taxonomic analysis showed for example that H. felis, H. salomonis and H. bizzozeronii could not be differentiated on the basis of the 16S rRNA gene sequence[28].
There are a number of genes which are theoretically specific for H pylori, eg ureA, glmM, and vacA. Although they were looked for in this study, they were not uniformly present. The absence of the H pylori specific vacA detected by a PCR targeting a short sequence is particularly surprising. Furthermore, a striking feature in the 16S rDNA sequences obtained from liver material is the presence of 2 nucleotide polymorphisms at the positions 92 and 130 as described by Verhoef et al[29]. Indeed, this polymorphism was already present in the sequences first described by Avenaud et al[8] and Ponzetto et al[11], and was present in most of our cases (5/7). It is important to note that in the study of Verhoef et al[29], H pylori was grown from the stomach of 3 out of 9 (33%) patients with this Helicobacter 16S rDNA polymorphism in the liver, and the corresponding gastric strains also had this polymorphism and were confirmed to be H pylori. It is therefore most likely that this species corresponds to a variant of H pylori with different properties including bile resistance and the ability to colonize the liver. This polymorphism is also present in the only strain grown from liver material[10]. The study of strains obtained from the same patients several years apart has shown a greater diversity than in other bacterial species studied due to high rates of mutations and recombinations[30]. This observation has led to the concept of quasi species which could correspond to the situation found in the liver.
Despite the doubt concerning the reality of the presence of H pylori in the liver (absence of culture and negative-H pylori specific PCR), the presence of H. spullorum-like organisms in the liver is much more likely as has been confirmed by H. spullorum specific cdtB PCR and by sequencing of the 16S rRNA gene. The identification of H. pullorum is easy, given that this bacterium has been designated as a separate species on the basis of 16S rRNA gene sequencing[31]. H. pullorum isolates were initially recovered from the cecal content of broiler chickens and from the liver and intestinal content of laying hens with vibrionic hepatitis, suggesting that this bacterium can infect the liver and that poultry may serve as the source of human infection[31]. H. pullorum has also been cultured from immunocompetent patients with gastroenteritis and one HIV-infected patient[31]. One individual, in addition to having diarrhea, developed elevated liver enzyme levels and hepatomegaly which, although not proven, may have been induced by H. pullorum invasion of the liver in a manner similar to the organism’s ability to cause hepatitis in chickens[32]. There is clearly a potential for zoonotic food-borne transmission of H. pullorum to humans, as is known to occur with Campylobacter species.
In conclusion, Helicobacter DNA can be present in the liver of patients with liver disease, with a tendency to have a higher prevalence in those with cirrhosis. Although H. pullorum appears to be regularly found at a low rate, the exact nature of the main Helicobacter species present is still uncertain. It is most likely a variant of H pylori which has acquired specific properties. Further studies exploring both the liver and stomach of patients with liver diseases need to be carried out to unravel the mystery of this intriguing association.
The authors thank the ANRS and the Conseil Régional d’Aquitaine for supporting the project.
| 1. | Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345:41-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2042] [Cited by in RCA: 2026] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 2. | Marcellin P, Asselah T, Boyer N. Fibrosis and disease progression in hepatitis C. Hepatology. 2002;36:S47-S56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 151] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 3. | Poynard T, Bedossa P, Opolon P. Natural history of liver fibrosis progression in patients with chronic hepatitis C. The OBSVIRC, METAVIR, CLINIVIR, and DOSVIRC groups. Lancet. 1997;349:825-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2199] [Cited by in RCA: 2168] [Article Influence: 74.8] [Reference Citation Analysis (0)] |
| 4. | Poynard T, Ratziu V, Charlotte F, Goodman Z, McHutchison J, Albrecht J. Rates and risk factors of liver fibrosis progression in patients with chronic hepatitis c. J Hepatol. 2001;34:730-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 501] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 5. | Roudot-Thoraval F, Bastie A, Pawlotsky JM, Dhumeaux D. Epidemiological factors affecting the severity of hepatitis C virus-related liver disease: a French survey of 6,664 patients. The Study Group for the Prevalence and the Epidemiology of Hepatitis C Virus. Hepatology. 1997;26:485-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 195] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 6. | Castéra L, Hézode C, Roudot-Thoraval F, Bastie A, Zafrani ES, Pawlotsky JM, Dhumeaux D. Worsening of steatosis is an independent factor of fibrosis progression in untreated patients with chronic hepatitis C and paired liver biopsies. Gut. 2003;52:288-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 214] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 7. | Ward JM, Fox JG, Anver MR, Haines DC, George CV, Collins MJ Jr, Gorelick PL, Nagashima K, Gonda MA, Gilden RV. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst. 1994;86:1222-1227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 278] [Article Influence: 8.7] [Reference Citation Analysis (1)] |
| 8. | Avenaud P, Marais A, Monteiro L, Le Bail B, Bioulac Sage P, Balabaud C, Mégraud F. Detection of Helicobacter species in the liver of patients with and without primary liver carcinoma. Cancer. 2000;89:1431-1439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 9. | Solnick JV, Schauer DB. Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin Microbiol Rev. 2001;14:59-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 263] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 10. | de Magalhães Queiroz DM, Santos A. Isolation of a Helicobacter strain from the human liver. Gastroenterology. 2001;121:1023-1024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 11. | Ponzetto A, Pellicano R, Leone N, Cutufia MA, Turrini F, Grigioni WF, D'Errico A, Mortimer P, Rizzetto M, Silengo L. Helicobacter infection and cirrhosis in hepatitis C virus carriage: is it an innocent bystander or a troublemaker. Med Hypotheses. 2000;54:275-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 60] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 12. | Rocha M, Avenaud P, Ménard A, Le Bail B, Balabaud C, Bioulac-Sage P, de Magalhães Queiroz DM, Mégraud F. Association of Helicobacter species with hepatitis C cirrhosis with or without hepatocellular carcinoma. Gut. 2005;54:396-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 120] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 13. | Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2860] [Cited by in RCA: 3128] [Article Influence: 104.3] [Reference Citation Analysis (1)] |
| 14. | Bedossa P; IARC. Tumors of the liver and intrahepatic bile ducts. In: Pathology and genetics of tumours of the digestive system. Lyon 2000; 157-217. |
| 15. | Monteiro L, de Mascarel A, Sarrasqueta AM, Bergey B, Barberis C, Talby P, Roux D, Shouler L, Goldfain D, Lamouliatte H. Diagnosis of Helicobacter pylori infection: noninvasive methods compared to invasive methods and evaluation of two new tests. Am J Gastroenterol. 2001;96:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 125] [Article Influence: 5.0] [Reference Citation Analysis (1)] |
| 16. | Mégraud F, Lehn N, Lind T, Bayerdörffer E, O'Morain C, Spiller R, Unge P, van Zanten SV, Wrangstadh M, Burman CF. Antimicrobial susceptibility testing of Helicobacter pylori in a large multicenter trial: the MACH 2 study. Antimicrob Agents Chemother. 1999;43:2747-2752. [PubMed] |
| 17. | Fox JG, Dewhirst FE, Shen Z, Feng Y, Taylor NS, Paster BJ, Ericson RL, Lau CN, Correa P, Araya JC. Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology. 1998;114:755-763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 345] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 18. | Germani Y, Dauga C, Duval P, Huerre M, Levy M, Pialoux G, Sansonetti P, Grimont PA. Strategy for the detection of Helicobacter species by amplification of 16S rRNA genes and identification of H. felis in a human gastric biopsy. Res Microbiol. 1997;148:315-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 78] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 19. | Oleastro M, Ménard A, Santos A, Lamouliatte H, Monteiro L, Barthélémy P, Mégraud F. Real-time PCR assay for rapid and accurate detection of point mutations conferring resistance to clarithromycin in Helicobacter pylori. J Clin Microbiol. 2003;41:397-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 163] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 20. | Boonjakuakul JK, Syvanen M, Suryaprasad A, Bowlus CL, Solnick JV. Transcription profile of Helicobacter pylori in the human stomach reflects its physiology in vivo. J Infect Dis. 2004;190:946-956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 21. | He Q, Wang JP, Osato M, Lachman LB. Real-time quantitative PCR for detection of Helicobacter pylori. J Clin Microbiol. 2002;40:3720-3728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 89] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 22. | Mikula M, Dzwonek A, Jagusztyn-Krynicka K, Ostrowski J. Quantitative detection for low levels of Helicobacter pylori infection in experimentally infected mice by real-time PCR. J Microbiol Methods. 2003;55:351-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 23. | Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389-3402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53569] [Cited by in RCA: 52905] [Article Influence: 1824.3] [Reference Citation Analysis (0)] |
| 24. | Huang Y, Fan XG, Wang ZM, Zhou JH, Tian XF, Li N. Identification of helicobacter species in human liver samples from patients with primary hepatocellular carcinoma. J Clin Pathol. 2004;57:1273-1277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 121] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 25. | Oliveira AG, das Graças Pimenta Sanna M, Rocha GA, Rocha AM, Santos A, Dani R, Marinho FP, Moreira LS, de Lourdes Abreu Ferrari M, Moura SB. Helicobacter species in the intestinal mucosa of patients with ulcerative colitis. J Clin Microbiol. 2004;42:384-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 26. | Oliveira AG, Rocha GA, Rocha AM, Sanna Md, Moura SB, Dani R, Marinho FP, Moreira LS, Ferrari Mde L, Castro LP. Isolation of Helicobacter pylori from the intestinal mucosa of patients with Crohn's disease. Helicobacter. 2006;11:2-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 27. | Feldman RA, Deeks JJ, Evans SJ. Multi-laboratory comparison of eight commercially available Helicobacter pylori serology kits. Helicobacter pylori Serology Study Group. Eur J Clin Microbiol Infect Dis. 1995;14:428-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 72] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | O'Rourke JL, Solnick JV, Neilan BA, Seidel K, Hayter R, Hansen LM, Lee A. Description of 'Candidatus Helicobacter heilmannii' based on DNA sequence analysis of 16S rRNA and urease genes. Int J Syst Evol Microbiol. 2004;54:2203-2211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 105] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 29. | Verhoef C, Pot RG, de Man RA, Zondervan PE, Kuipers EJ, IJzermans JN, Kusters JG. Detection of identical Helicobacter DNA in the stomach and in the non-cirrhotic liver of patients with hepatocellular carcinoma. Eur J Gastroenterol Hepatol. 2003;15:1171-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 44] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 30. | Kuipers EJ, Israel DA, Kusters JG, Gerrits MM, Weel J, van Der Ende A, van Der Hulst RW, Wirth HP, Höök-Nikanne J, Thompson SA. Quasispecies development of Helicobacter pylori observed in paired isolates obtained years apart from the same host. J Infect Dis. 2000;181:273-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 134] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 31. | Stanley J, Linton D, Burnens AP, Dewhirst FE, On SL, Porter A, Owen RJ, Costas M. Helicobacter pullorum sp. nov.-genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology. 1994;140:3441-3449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 159] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 32. | Burnens AP, Stanley J, Morgenstern R, Nicolet J. Gastroenteritis associated with Helicobacter pullorum. Lancet. 1994;344:1569-1570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 59] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
S- Editor Wang GP L- Editor Wang XL E- Editor Bai SH