Published online Nov 28, 2006. doi: 10.3748/wjg.v12.i44.7113
Revised: August 28, 2006
Accepted: September 18, 2006
Published online: November 28, 2006
AIM: To investigate the functions of promoter hyper-methylation of secreted frizzled-related proteins (sFRPs) genes in colorectal tumorigenesis and progression.
METHODS: The promoter hypermethylation and expression of sFRP genes in 72 sporadic colorectal carcinomas, 33 adenomas, 18 aberrant crypt foci (ACF) and colorectal cancer cell lines RKO, HCT116 and SW480 were detected by methylation-specific PCR and reverse transcription PCR, respectively.
RESULTS: None of the normal colorectal mucosa tissues showed methylated bands of any of four sFRP genes. sFRP1, 2, 4 and 5 were frequently methylated in colorectal carcinoma, adenoma and ACF (sFRP1 > 85%, sFRP2 >75%, sFRP5 > 50%), and the differences between three colorectal tissues were not significant (P > 0.05). Methylation in colorectal tumors was more frequent than in normal mucosa and adjacent normal mucosa. The mRNA of sFRP1-5 genes was expressed in all normal colorectal mucosa samples. Expression of sFRP1, 2, 4 and 5 and sFRP1, 2 and 5 was downregulated in carcinoma and adenoma, respectively. The downregulation of sFRP2, 4 and 5 was more frequent in carcinoma than in adenoma. Expression of sFRP3 which promoter has no CpG island was downregulated in only a few of colorectal tumor samples (7/105). The downregulation of sFRP1, 2, 4 and 5 expression was significantly associated with promoter hypermethylation in colorectal tumor. After cells were treated by DAC/TSA combination, the silenced sFRP mRNA expression could be effectively re-expressed in colorectal cancer cell lines.
CONCLUSION: Hypermethylation of sFRP genes is a common early event in the evolution of colorectal tumor, occurring frequently in ACF, which is regarded as the earliest lesion of multistage colorectal carcinogenesis. It appears to functionally silence sFRP genes expression. Methylation of sFRP1, 2 and 5 genes might serve as indicators for colorectal tumor.
- Citation: Qi J, Zhu YQ, Luo J, Tao WH. Hypermethylation and expression regulation of secreted frizzled-related protein genes in colorectal tumor. World J Gastroenterol 2006; 12(44): 7113-7117
- URL: https://www.wjgnet.com/1007-9327/full/v12/i44/7113.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i44.7113
Secreted frizzled-related proteins (sFRPs) comprise a family of five secreted glycoproteins that antagonize Wnt canonical and noncanonical signaling by different mechanisms directly or indirectly[1]. Wnt signaling regulates cell growth, motility and differentiation in animal development and has increasingly been implicated in tissue homeostasis in adult organisms. Aberrant Wnt signaling pathway is an early progression event in 90% of colorectal cancers, contributing to the growth, proliferation and loss of apoptosis of tumor cells[2]. Thus, role of sFRP as a negative regulator of Wnt signaling may have important implications in tumorigenesis, and its downregulation has been correlated with human cancers. To date, no mutation in sFRP genes has been associated with their dysregulation in tumors. Aberrant hypermethylation of CpG islands in gene promoter has been found to be a primary mechanism in the inactivation of several tumor suppressor genes. As we know, colorectal tumor is one of the tumors with high frequencies of gene methylation[3,4]. Epigenetic gene silencing plays an extremely important role in the early stage of colorectal tumorigenesis. In this study, we examined hypermethylation and expression of sFRP genes in different stages of colorectal tumor and colorectal cancer cell lines in order to investigate the functions of promoter hypermethylation of sFRP genes in colorectal tumorigenesis and progression, and whether it can serve as an indicator for colorectal tumor.
Tissue samples: Tissue samples, including 72 sporadic colorectal carcinomas, 33 sporadic colorectal adenomas (male 56, female 49, age 27-76 years), 38 adjacent normal mucosa from 28 colorectal carcinomas and 15 colorectal adenomas, and 20 normal colorectal mucosa tissues (male 11, female 9, age 20-54 years) were obtained from surgical resection of patients receiving endoscopy examination at the clinic of Zhongnan Hospital of Wuhan University and The Tumor Hospital of Hubei Province between October 2003 and April 2006. Eighteen ACFs from 15 colorectal carcinomas were also collected[5]. Samples were stored at -70°C until processing. None of the patients had received chemotherapy or radiation therapy prior to surgery. All patients gave informed consent for their participation in the study which had been approved by the Ethical Committee of our university.
Cell culture and treatment: Three colorectal carcinoma cell lines, including RKO, HCT116, and SW480 (cell lines were purchased from China Center for Type Culture Collection) were used in this study. Cell lines were cultured in RPMI 1640 medium (GIBCO BRL) supplemented with 10% fetal bovine serum and treated with 5-aza-2’-deoxycytidine (DAC), a DNA methyltransferase (DNMT) inhibitor, and trichostatin A (TSA), a histone deacetylase (HDAC) inhibitor. Three cell lines all received four different treatments: low-dose DAC (200 nmol/L, 48 h), high-dose DAC (5 μmol/L, 72 h), TSA (300 nmol/L from a 1.5 mmol/L ethanol-dissolved stock, 24 h), and DAC (200 nmol/L, 48 h) followed by addition of TSA (300 nmol/L, 24 h), the drugs and medium being replaced every 24 h.
Methylation analysis: Methylation-specific PCR (MSP) was used to examine the promoter hypermethylation of sFRP1, 2, 4 and 5 genes. Genomic DNA was extracted by the phenol-chloroform standard methods. Bisulfite genomic DNA modification and purification was performed as previously reported[6]. The modified DNA was amplified by methylated and unmethylated primers (exact primers are available upon request). Products were visualized by 12% polyacrylamide gel electrophoresis. MSP was not performed for sFRP3, which has no CpG island in promoter region.
Reverse transcription PCR (RT-PCR) analysis: RT-PCR was employed to detect sFRP1-5 genes mRNA expression. TRIZOL Reagent (Invitrogen) was used to extract total RNA from tissues and cells. The integrity of RNA was identified by methanal denatured agarose electrophoresis. Ultraviolet-visible light spectrophotometer (Beckman Coulter DU530) was used to determine A260/280 of total RNA. Reverse transcription was performed on 1 μg of total RNA in a reaction volume of 20 μL. PCR assays were performed in 50 μL volume using 100 ng cDNA. Glyceraldehyde phosphate dehydrogenase (GAPDH) was employed as an internal reference gene to ensure cDNA quality and loading accuracy. Products were visualized by 2.5% agarose electrophoresis, and then analyzed by gel imaging system (SynGene GGM) and GeneTools software. RT-PCR was performed three times for the tumor samples with detectable mRNA, and then the gene expression index (density Lum of samples mRNA/density Lum of GAPDH mRNA) of these samples was compared with the normal samples (t test). Those with reduced mRNA expression and P < 0.01 were considered as significantly downregulated.
Statistical analysis was performed using SPSS 11.5 software. Associations between the discrete variables were assessed using the two-sided Fisher’s exact test or Pearson’s chi square tests. P value less than 0.05 was regarded as statistically significant.
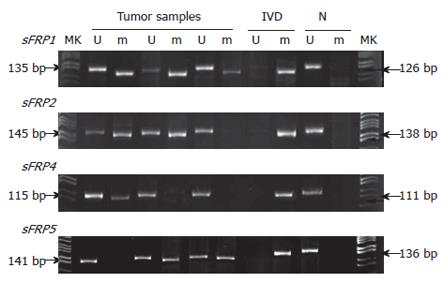
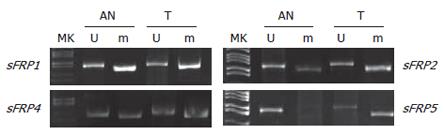
None of normal mucosa showed methylated bands. SFRP1, 2, 4 and 5 were methylated in 93.1% (67/72), 83.3% (60/72), 36.1% (26/72) and 52.8% (38/72) of colorectal carcinoma; 87.9% (29/33), 81.8% (27/33), 24.2% (8/33) and 57.6% (19/33) of colorectal adenoma (Figure 1); 52.6% (20/38), 28.9% (11/38), 2.6% (1/38) and 18.4% (7/38) of adjacent normal mucosa to colorectal carcinomas and adenomas, respectively. Most of sFRP genes were methylated in tumor samples when methylation was presented in the adjacent normal mucosa from the same sample, sFRP1 (20/20), sFRP2 (9/11), sFRP4 (1/1) and sFRP5 (6/7). The tumors showed stronger methylation signals than the adjacent normal tissues (Figure 2).
The methylation of each of the sFRP1, 2, 4 and 5 genes was significantly different between carcinoma and normal mucosa (P < 0.001 for sFRP1, 2 and 5, P = 0.002 for sFRP4), between carcinoma and adjacent normal mucosa (P < 0.001 for sFRP1, 2, 4 and 5), between adenoma and normal mucosa (P < 0.001 for sFRP1, 2 and 5, P = 0.0462 for sFRP4), and between adenoma and adjacent normal mucosa (P = 0.001 for sFRP1, P < 0.001 for sFRP2 and 5, P = 0.018 for sFRP4). While no significant difference was found between carcinoma and adenoma (P = 0.614 for sFRP1, P = 0.848 for sFRP2, P = 0.228 for sFRP4 and P = 0.647 for sFRP5).
SFRP1, 2, 4 and 5 were methylated in 94.4% (17/18), 77.8% (14/18), 27.8% (5/18) and 55.6% (10/18) of 18 ACF samples. SFRP1, 2, 4 and 5 methylation differed significantly between ACF and normal mucosa (P < 0.001 for sFRP1, 2 and 5 and P = 0.017 for sFRP4, Fisher’s exact test). While no significant difference was found between ACF and carcinoma (P = 1.000 for sFRP1, P = 0.836 for sFRP2, P = 0.506 for sFRP4 and P = 0.833 for sFRP5), and between ACF and adenoma (P = 0.794 for sFRP1, P = 1.000 for sFRP2, P = 1.000 for sFRP4 and P = 0.059 for sFRP5). At least one of the four sFRP genes was methylated in 18 ACF samples.
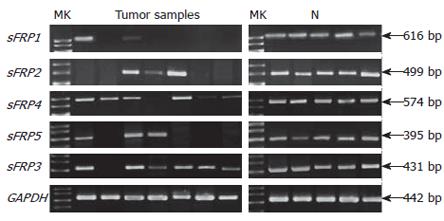
The 20 normal colorectal mucosa specimens expressed considerable levels of sFRP1-5. Compared with the normal mucosa, sFRP1, 2, 4 and 5 expression was silenced or significantly reduced in 90.3% (65/72), 70.8% (51/72), 26.4% (19/72) and 61.1% (44/72) of colorectal carcinomas, respectively (P < 0.001 for sFRP1, 2 and 5 and P = 0.023 for sFRP4); sFRP1, 2, and 5 were markedly downregulated or silenced in 75.8% (25/33), 45.5% (15/33) and 39.4% (13/33) of colorectal adenomas (P < 0.001 for sFRP1, 2 and 5). SFRP4 was downregulated or silenced in 9.1% (3/33) of colorectal adenomas relative to the normal mucosa (P = 0.438). The downregulation of sFRP 2, 4 and 5 expression was more frequent in carcinoma than in adenoma (P = 0.012, P = 0.043 and P = 0.038). SFRP1 expression had no significant difference between carcinoma and adenoma (P = 0.094). A minority of adjacent normal mucosa samples had reduced expression of sFRP1-5 genes, and the ratio was 13.2% (5/38), 5.3% (2/38), 0% (0/38), 7.9% (3/38) and 7.9% (3/38), respectively. SFRP3 that has no CpG island in promoter region was downregulated in only 7 of 105 colorectal tumor samples (Figure 3). Reduced expression of sFRP1, 2, 4 and 5 genes was significantly associated with aberrant hypermethylation of these genes (Table 1).
| Gene | Colorectal tumor | |||
| Unmethylated | Methylated | χ2, P | ||
| sFRP1 | + | 6 | 9 | χ2 = 17.627 |
| - | 3 | 87 | P < 0.001 | |
| sFRP2 | + | 11 | 28 | χ2 = 5.345 |
| - | 7 | 59 | P = 0.021 | |
| sFRP4 | + | 68 | 15 | χ2 = 37.041 |
| - | 3 | 19 | P < 0.001 | |
| sFRP5 | + | 38 | 10 | χ2 = 39.872 |
| - | 10 | 47 | P < 0.001 | |
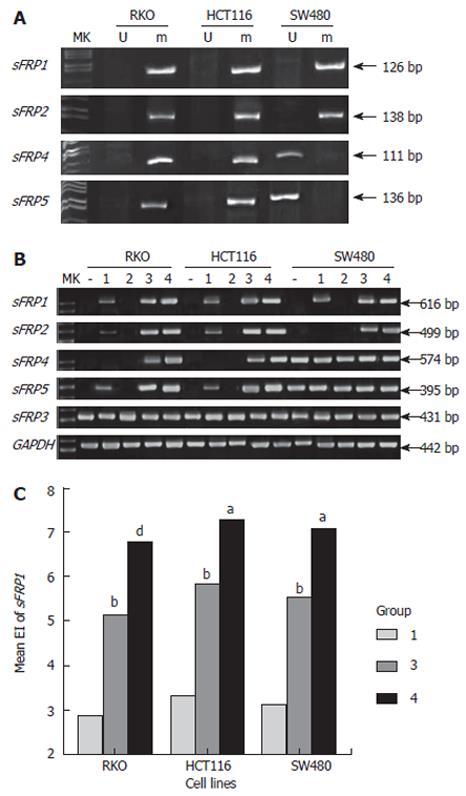
SFRP1, 2, 4 and 5 genes were methylated in RKO and HCT116 cell lines. SFRP1 and 2 but not sFRP 4 and 5 were also methylated in SW480 cell line (Figure 4A). When sFRP genes were methylated, their corresponding mRNA expression was absent in three colorectal carcinoma cell lines. SW480 cell line expressed sFRP 4 and 5 mRNA, in which methylation was not detected in sFRP 4 and 5 promoter. SFRP3 was expressed in three colorectal carcinoma cell lines.
However, after cells were treated by low-dose DAC, the silenced sFRP1, 2 and 5 mRNA expression could be re-expressed in RKO and HCT116 cell lines; and sFRP1 expression could be re-expressed in SW480 cell line. High-dose DAC treatment could strengthen the effects of low-dose DAC treatment, and result in reexpression of sFRP 4 (RKO and HCT116) and 2 (SW480) mRNA, which could not be induced by low-dose DAC. The addition of TSA to DAC rendered the silenced sFRP genes mRNA to be re-expressed effectively, and markedly elevated the mRNA expression level that had been re-expressed or increased by high-dose DAC. A single application of TSA could not induce re-expression of sFRP genes mRNA. The influence of demethylation treatment on sFRP3 expression was minimal (Figure 4B and 4C).
Aberrant activation of the Wnt canonical signaling pathway is associated with a variety of human cancers, such as head and neck carcinoma, lung cancer, melanoma, and mesothelioma, and particularly colorectal cancer, as well as the growth, proliferation and loss of apoptosis of tumor cells. The tumor suppressor adenomatous polyposis coli (APC) mutates in a high proportion of colorectal carcinomas; meanwhile it is an important member of Wnt canonical pathway involved in the degradation of β-catenin. Wnt signaling has been identified as one of the key signaling pathways in cancer, regulating cell growth, motility and differentiation. It has been reported that the noncanonical pathway is also involved in tumorigenesis, being associated with inhibition of apoptosis[7].
SFRPs contain a characteristic cysteine-rich domain (CRD) that shares homology with the CRD of frizzled (Fz) receptor of Wnt in the N-terminal half of the proteins. Thus, sFRPs may block Wnt signaling either by interacting with Wnt proteins to prevent them from binding to Fz proteins or by forming nonfunctional complexes with Fz[1]. It has been reported that sFRPs can potentially inhibit the entire canonical Wnt pathway even in the presence of activating mutations of APC or β-catenin downstream of the Fz receptor in colon cancer cell[8]. Therefore, silence of sFRP genes may be essential for the aberrant activation of the Wnt canonical pathway in colorectal tumorigenesis.
To date, no mutation in sFRP genes has been associated with their dysregulation in tumors. Aberrant hypermethylation of CpG islands in gene promoters has been found to be a primary mechanism in the inactivation of several tumor suppressor genes. However, the presence of gene silencing by hypermethylation is an important character of colorectal tumor[3,4]. This study demonstrated that genes of the sFRP family were methylated in most cases of colorectal carcinoma, adenoma and ACF, especially sFRP1 and 2. The proportion of sFRP1 and 2 methylation was more than 85% and 75%, respectively. However, sFRP genes methylation was much less common in normal-appearing epithelia adjacent to colorectal tumor and never detected in normal mucosa. These data suggest that methylation of sFRP genes occurs as an early event in the evolution of ACF-adenoma-carcinoma sequence and is increased through carcinogenic transformation. Notably, 80.6% of colorectal carcinomas showed methylation of both sFRP1 and 2, and at least one of the four sFRP genes with CpG islands was methylated in 97.2% of the colorectal carcinomas and all ACF samples. SFRP1, 2 and 5 appeared to be more specific for colorectal carcinoma, adenoma and ACF, and may be more suitable candidate markers for these lesions.
Based on our study, methylation represents a likely mechanism of sFRP gene silencing in colorectal tumor: (1) sFRP expression is absent or markedly decreased in colorectal carcinoma and adenoma but generally high in normal mucosa, which is associated with sFRP gene methylation; (2) mRNA expression of sFRP genes is absent in cancer cell lines with corresponding sFRP gene methylation but high in cell lines with unmethylated sFRP genes; and (3) sFRP gene expression is reactivated in methylated cancer cell lines treated by demethylation; (4) sFRP3 that has no CpG island in promoter region is expressed in cancer cell lines and a majority of tumor and normal samples. These data suggest sFRP gene silencing induced by promoter hypermethylation plays a key role in colorectal tumorigenesis by permitting aberrant activation of Wnt canonical signaling. Furthermore, we propose that the inhibiting effect of sFRP3 on Wnt signaling is weak. Although sFRP3 is still expressed in tumor cells, it is not sufficient to block Wnt signaling. To confirm that, further studies are needed on the action of Wnt signaling in colorectal tumor cells.
We also found promoter hypermethylation of sFRP genes was present at equal frequency in colorectal carcinoma and adenoma, but the downregulation of sFRP 2, 4 and 5 expression was more frequent in carcinoma than in adenoma. These suggest that sFRP genes are not completely silenced by promoter hypermethylation in the stage of adenoma, although methylation has been presented in adenoma and even early stage ACF. The silence of sFRP genes tends to increase with the colorectal tumor progression. Thus, the downregulation of sFRP genes may be associated with the progression and malignant potential of colorectal tumor. Our data showed sFRP1 methylation and downregulation were both extremely frequent in colorectal carcinomas and adenomas. It suggests the methylation and downregulation of sFRP1 occur more frequently and earlier than other sFRP genes in colorectal tumor, and perhaps play more important role in the progression of colorectal tumorigenesis. However, the methylation and downregulation of sFRP4 were less common than sFRP1, 2 and 5 genes in colorectal tumor, though they were both high in mesothelioma and esophageal adenocarcinoma as previously reported[9,10]. It has been reported sFRP4 has the least homology with other family members[11]. These suggest the Wnt signaling is regulated by different sFRP molecules in different histiocytes. In colorectal tumor cells, the role of sFRP1, 2 and 5, especially the sFRP1, is more important than sFRP3 and 4.
Two key epigenetic pathways are involved in gene transcription regulation: DNA methylation and histone acetylation. DNA methylation is carried out by three DNA methyltransferases (DNMT), DNMT1, DNMT3A, and DNMT3B. DNA methylation represses transcription directly, by inhibiting the binding of specific transcription factors, and indirectly, by recruiting methyl-CpG-binding proteins and their associated repressive chromatin remodeling activities[12]. Deacetylation catalyzed by histone deacetylases is important for turning off genes and maintaining some genes in a repressed state[13]. DAC is a DNMT inhibitor that induces DNA demethylation by metabolic incorporation into genomic DNA, thereby covalently arresting DNMT. TSA is an HDAC inhibitor that causes hyperacetylation on histones H3 and H4. Our data showed sFRP genes were effectively demethylated by the high-dose DAC or combination of DAC and TSA treatment and re-expressed in colorectal cancer cell lines. The effect of combination of DAC and TSA treatment was better than high-dose DAC, even though the DAC concentration was low in the combination. A single application of TSA could not induce re-expression of sFRP genes mRNA, although some genes were upregulated in expression by TSA alone in the previous report[14]. These data indicate DNA methylation and histone deacetylation are synergetic in the inhibition of gene transcription, and in terms of sFRP genes, DNA methylation plays the dominant role.
Our study shows that hypermethylation of sFRP genes is a common early event in the evolution of colorectal tumor, and hypermethylation patterns of sFRP genes may provide a potentially useful marker system for predicting the risk of colonic neoplasia.
Silence of sFRP genes induced by promoter hyperme-thylation plays a key role in colorectal tumorigenesis. As DNA sequence is not altered by epigenetic modification, and gene silenced by promoter hypermethylation could normally transcript after demethylation, modulation of Wnt protein-driven cell growth, through reversal of sFRP genes silencing, may represent potential targets for colorectal cancer prevention and treatment.
We thank Dr. Bing Xiong, Qun Qian and Liao-Bing Chen for providing and storing CRC and normal samples, and Jun-Zhu Wu for helpful comments on the manuscript.
| 1. | Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627-2634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1193] [Cited by in RCA: 1261] [Article Influence: 54.8] [Reference Citation Analysis (1)] |
| 2. | Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001;1:55-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 659] [Cited by in RCA: 673] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 3. | Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225-3229. [PubMed] |
| 4. | Qi J, Zhu YQ, Huang MF, Yang D. Hypermethylation of CpG island in O6-methylguanine-DNA methyltransferase gene was associated with K-ras G to A mutation in colorectal tumor. World J Gastroenterol. 2005;11:2022-2025. [PubMed] |
| 5. | Bouzourene H, Chaubert P, Seelentag W, Bosman FT, Saraga E. Aberrant crypt foci in patients with neoplastic and nonneoplastic colonic disease. Hum Pathol. 1999;30:66-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 50] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 6. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4183] [Cited by in RCA: 4262] [Article Influence: 142.1] [Reference Citation Analysis (12)] |
| 7. | Doucas H, Garcea G, Neal CP, Manson MM, Berry DP. Changes in the Wnt signalling pathway in gastrointestinal cancers and their prognostic significance. Eur J Cancer. 2005;41:365-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 57] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 8. | Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 821] [Cited by in RCA: 844] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 9. | Lee AY, He B, You L, Dadfarmay S, Xu Z, Mazieres J, Mikami I, McCormick F, Jablons DM. Expression of the secreted frizzled-related protein gene family is downregulated in human mesothelioma. Oncogene. 2004;23:6672-6676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 158] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 10. | Zou H, Molina JR, Harrington JJ, Osborn NK, Klatt KK, Romero Y, Burgart LJ, Ahlquist DA. Aberrant methylation of secreted frizzled-related protein genes in esophageal adenocarcinoma and Barrett's esophagus. Int J Cancer. 2005;116:584-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 124] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 11. | Jones SE, Jomary C. Secreted Frizzled-related proteins: searching for relationships and patterns. Bioessays. 2002;24:811-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 324] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 12. | Turek-Plewa J, Jagodziński PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631-647. [PubMed] |
| 13. | LaVoie HA. Epigenetic control of ovarian function: the emerging role of histone modifications. Mol Cell Endocrinol. 2005;243:12-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 42] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 14. | Xiong Y, Dowdy SC, Podratz KC, Jin F, Attewell JR, Eberhardt NL, Jiang SW. Histone deacetylase inhibitors decrease DNA methyltransferase-3B messenger RNA stability and down-regulate de novo DNA methyltransferase activity in human endometrial cells. Cancer Res. 2005;65:2684-2689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 118] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
S- Editor Wang J L- Editor Zhu LH E- Editor Ma WH