Published online Nov 14, 2006. doi: 10.3748/wjg.v12.i42.6893
Revised: September 6, 2006
Accepted: September 12, 2006
Published online: November 14, 2006
AIM: To identify genes differentially expressed in mouse hepatocarcinoma ascites cell line with low potential of lymphogenous metastasis.
METHODS: A subtracted cDNA library of mouse hepatocarcinoma cell line with low potential of lympho-genous metastasis Hca-P and its synogenetic cell line Hca-F with high metastatic potential was constructed by suppression subtracted hybridization (SSH) method. The screened clones of the subtracted library were sequenced and GenBank homology search was performed.
RESULTS: Fifteen differentially expressed cDNA fragments of Hca-P were obtained which revealed 8 known genes, 4 expressed sequence tags (ESTs) and 3 cDNAs showed no homology.
CONCLUSION: Tumor metastasis is an incident involving multiple genes. SSH is a useful technique to detect differentially expressed genes and an effective method to clone novel genes.
- Citation: Cui XN, Tang JW, Hou L, Song B, Ban LY. Identification of differentially expressed genes in mouse hepatocarcinoma ascites cell line with low potential of lymphogenous metastasis. World J Gastroenterol 2006; 12(42): 6893-6897
- URL: https://www.wjgnet.com/1007-9327/full/v12/i42/6893.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i42.6893
Tumor metastasis is an incident involving multiple genes. However, the number of metastasis related genes available nowadays is very limited to elucidate the puzzling process of metastasis. Therefore, more attentions have been paid to screen candidate genes responsible for metastasis by high throughput technique. Hca-P and Hca-F are a pair of synogenetic mouse hepatocarcinoma ascites cell lines, possessing a specific potential of lymphogenous metastasis when inoculated subcutaneously into 615 mice, Hca-P showing a low metastatic potential (< 30%), while Hca-F showing a high potential (> 80%)[1]. In the current study, we employed suppressive subtracted hybridization (SSH) technique to identify differentially expressed genes specific for Hca-P in an effort to obtain candidate genes related to lymphogenous metastasis of hepatocarcinoma in mice.
Hca-F and Hca-P have been established and maintained by our laboratory[1]; inbred 615-mice were provided by the experimental animal center of our university.
Sixty inbred 615-mice were randomly divided into 2 groups. The Hca-P and Hca-F tumor cell lines were inoculated at 2 × 106 tumor cells of approximately 0.1 mL cell suspension into each mouse subcutaneously in each group. The mice were decapitated on the 28th day post-inoculation. The implanted tumor and the regional lymph nodes were removed and paraffin sections of tissues were HE stained and examined under microscope. The lymph node metastatic rates of Hca-F and Hca-P tumor cells were calculated.
Preparation of total RNA and mRNA: Isolation of total RNA was performed by TRIZOLTM(GIBCOBRL) and that of mRNA was carried out according to the protocol of oligotex mRNA spin column purification kit (Qiagen). The quantity and integrity of mRNA were detected by ultraviolet spectrometer and by electrophoresis on a denaturing formaldehyde agarose stained by EB. mRNA of Hca-P served as tester and mRNA of Hca-F as driver. SSH was performed between tester and driver by a PCR selectTM cDNA subtraction kit and 50 × PCR enzyme kit (Clontech, Heidelberg, Germany) following the instructions of the manufacturer.
dscDNA synthesis and digestion with RsaI: Briefly, 2 μg aliquots of each of poly (A+) mRNA from the tester and the pooled driver were subjected to dscDNA synthesis. Thereafter, they were purified by passing through Chroma spin-400 columns (Clontech, USA). Each purified dscDNA was digested with RsaI.
ligation to adaptor 1 and 2R: The tester cDNAs were subdivided into 2 equal groups and then ligated to adaptor 1 and 2R in separate ligation reactions. Ligation efficiency analysis was performed by amplifying ligation products with G3PDH 3’ primer/PCR primer 1 and G3PDH 3’ primer/G3PDH 5’ primer, respectively, and their intensity was compared.
Subtractive hybridization: Subtractive hybridization was performed by annealing an excess of driver cDNAs with each sample of adaptor-ligated tester cDNAs. The cDNAs were heat-denatured and incubated at 68°C for 8 h. After the first hybridization, the 2 samples were mixed together and hybridized again with freshly heat-denatured driver cDNAs for 20 h at 68°C. Two rounds of hybridization would generate a normalized population of tester-specific cDNAs with different adaptors at each end. After filling in the ends, 2 rounds of PCR amplification were performed to enrich desired cDNAs containing both adaptors by exponential amplification of these products[2]. The optimized cycles for the first and second PCRs were 27 and 13 respectively to increase representation and reduce redundancy of subtracted cDNA libraries.
Analysis of subtractive efficiency: Secondary PCR products were used as templates for PCR amplification of human G3PDH for 18, 23, 28 and 33 cycles respectively to assure subtraction efficiency. PCR products were run on 1.8% agarose gel.
Products of the secondary PCR reactions were cloned into a pT Adv vector (Clontech) and the resultant ligation products were then transformed into DH5α E. coli competent cells. The bacteria were subsequently grown in 800 μL of liquid Luria-Bertani medium and allowed to incubate for 45 min at 37°C with shaking at 150 rpm. Thereafter, the cells were plated onto agar plates containing ampicillin (50 μg/mL), 5-bromo-4-chloro-3-indoly-b-D-galactoside (X-gal; 20 μg/cm2) and iso-ploprl-b-D-thiogalactoside (IPTG; 12.1 μg/cm2) and incubated overnight at 37°C. Individual recombinant white clones were picked and grown in single line pattern onto Luria-Bertani agar solid medium containing ampicillin and allowed to incubate at 37°C for 6-7 h before single clone was picked from single-line pattern agar medium and allowed to grow in Luria-Bertani liquid medium containing ampicillin overnight at 37°C with shaking at 150 r/min.
Plasmids of candidate positive clones from subtracted cDNA library were isolated and amplified by PCR with nested primer 1 and primer 2. Meanwhile the product of PCR was detected by agarose gel electrophoresis.
Randomly screened 14 positive clones from the subtracted cDNA library were sequenced by T7/SP6 chain termination reaction in TaKaRa (DaLian, China). Nucleic acid homology searches were subsequently performed at the National Center of Biotechnology Information (National Institutes of Health, Bethesda, Md., NCBI).
Implanted tumors of both Hca-P tumor-bearing mice and Hca-F tumor-bearing mice were palpable on 7th day post-inoculation. On the 28th day post-inoculation, 10% Hca-P cells bearing mice developed metastatic regional lymph nodes (3/30), while 80% Hca-F cells bearing mice developed metastatic regional lymph nodes (24/30).
The RNA samples electrophoresed on 1% agarose/EB gel exhibited 2 typical bands, corresponding to ribosomal 28s and 18s RNA, respectively, with a ratio of intensities > 2:1 and 1.9, ideal A260/A28 ratios of both samples obtained, indicating high integrity and purification of the total RNA we obtained. mRNA samples appeared as a smear with weak ribosomal RNA band: a high-quality mRNA was purified.
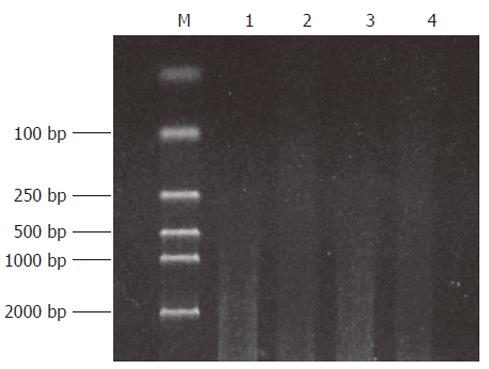
Both the digested cDNA and undigested cDNA usually presented as smears. However, their patterns were different. The digested cDNA fragments became shorter after RsaI digestion (Figure 1).
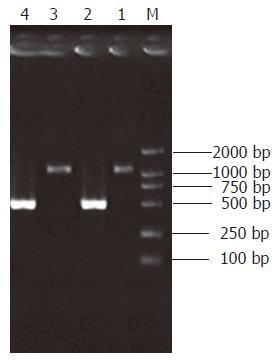
Intensity of the PCR product amplified using one gene-specific primer (G3PDH 3’ primer) and PCR primer 1 was 25% more than that of PCR product amplified using two gene-specific primers (G3PDH 3’ primer and 5’ primer). Ligation efficiency was > 25%, ensuring enough tester cDNA in the following hybridization (Figure 2).
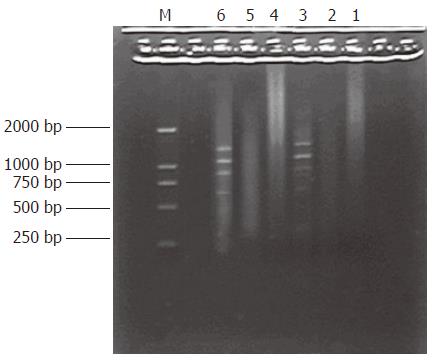
PCR products of the subtracted and unsubtracted usually looked like smears with or without discrete bands. However, the patterns between them were different
(Figure 3).
Subtraction efficiency analysis showed the effectively reduced amount of non-differentially expressed genes. In unsubtracted cDNA libraries, housekeeping gene G3PDH PCR products were visible after 23 cycles of amplification and became saturated after 23-28 cycles. However, subtracted libraries required 33 cycles for G3PDH to be detected (Figure 4).
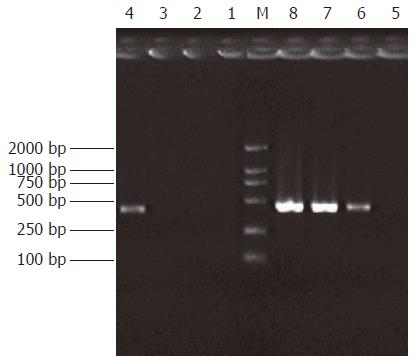
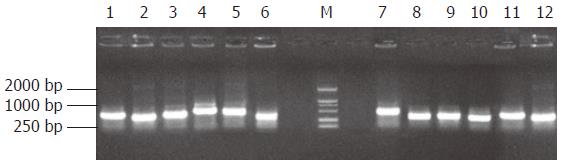
The subtracted cDNA libraries were composed of 995 positive clones, of which 200 clones were randomly picked up and plasmids of the candidate positive clones were isolated and amplified by PCR with nested primer 1 and primer 2. As a result, 189 positive clones showed PCR products of a size of 300-1000 bp (Figure 5).
Fifteen screened clones randomly selected were sequenced and homology search (http://www.ncbi.hlm.nih.gov/BLAST/) revealed 8 known genes and 4 expressed sequence tags (ESTs). Three cDNAs showed no ho-mology and presumably represented novel genes (Table 1).
| Clone seriel number | Size (bp) | Sequence identity |
| 1-3 | 508 | Mus musculus Telomere repeat binding factors TRF1 |
| 3-5 | 543 | Mus musculus Telomere repeat binding factors TRF2, |
| 9-6 | 489 | Mus musculus maspin |
| 15-4 | 335 | Mouse chromosome 3 clone RP 6-126M1 |
| 1-7 | 386 | Mouse 7 d embryo whole body cDNA RIKEN full-length enrich library, clone 2210102k3 |
| 16-3 | 503 | Mouse 5 d liver cells cDNA RIKEN full-length library enriched library, clone E330462F4 |
| 10-1 | 549 | Mouse chromosome 17 clone RP 26-122M3 |
| 13-7 | 502 | Mouse chromosome 4 clone RP13-110N10 |
| 9-8 | 340 | EST-mouse |
| 6-6 | 411 | EST-mouse |
| 11-2 | 390 | EST-mouse |
| 20-9 | 399 | EST-mouse |
| 23-3 | 470 | Unknown |
| 17-6 | 486 | Unknown |
| 12-3 | 344 | Unknown |
Tumor metastasis, as the leading cause of tumor related death, is a process involving multiple genes and their products. Elucidation of the gene expression profiles specific for tumor cells with different potential of metastasis might help in the understanding of the molecular mechanisms of metastasis. As one of the high throughput screening techniques, SSH technique has two distinct advantages: (1) it boasts a high subtraction efficiency; (2) it harbors an equalized representation of differentially expressed sequences which can separate effectively both high and low copy expressed genes mainly because of normalization[2]. von Stein et al[3] found about 94% positive rate in their research. Thus they considered confirmation of differentially expressed genes by Northern blot analysis for each clone obtained was probably unnecessary.
For a long time, studies have focused on the angiogenesis of tumors, but the roles of lymphatic vessels in tumor growth and metastasis were neglected. However, it is well known that lymphatic metastasis is mainly responsible for the spread of epithelial malignant tumors, and is closely related to the prognosis of patients.
Hca-P and Hca-F are a pair of synogenetic mouse hepatocarcinoma ascites cell lines presenting a specific potential of lymphogenous metastasis with a significant difference in their potential of metastasis[1]. Candidate genes involved in lymphogenous metastasis are supposed to be among the differentially expressed genes.
Using Hca-P as a tester, Hca-F as a driver, and we employed SSH technique to identify differentially expressed genes specific for Hca-P(low metastatic potential) so as to obtain candidate suppressor genes of lymphogenous metastasis. Fifteen screened clones randomly selected were sequenced and homology search revealed 8 known genes as TRF1, TRF2(telomere repeat binding factor 1, 2) ; maspin; mouse 7 days embryo whole body cDNA, RIKEN full-length enriched library clone 2210102k3; mouse 5 days liver cells cDNA, RIKEN full-length enriched library, clone E330462F4; mouse chromosome 3 clone RP 6-126M1;mouse chromosome 17 clone RP 26-122M3 and mouse chromosome 4 clone RP13-110N10. Studies showed TRF1, and TRF2 play important roles in genome stabilization [4-11] and are down-regulated in some malignant cell lines and tumor tissues[12-18]. In hematopoietic carcinogenesis, gene expression of telomerase suppressors such as TRF and TIN2, is decreased. mRNA encoding TRF1 and TRF2 when gastric cancer becomes more deeply invaded, is significantly decreased, indicating a negative association with tumor progression. Of the 8 known fragments, one showed high homology to mouse Maspin gene. Serving as one of the few p53-targeted genes involved in tumor invasion and metastasis, Maspin, a member of the serpin family, has been reported to suppress metastasis and angiogenesis in breast and prostate cancers, and is closely correlated with their clinical manifestations[19-31]. It indicates that SSH in our study is capable of enriching metastasis related genes.
Another 5 known fragments were attributed to embryo genes. Embryo genes AFP and CEA are overexpressed in hepatocarcinoma and other malignant tumors, indicating a possible association between embryo development and tumor. Embryo genes were also found in our previously established SSH library which contains candidate tumor metastasis genes[32]. These data showed that up-regulated expression of embryo genes during metastasis is not a casual event. Their roles in tumor metastasis need to be clarified. Moreover, 4 cDNA fragments demonstrated homology with 4 ESTs-mouse and 3 cDNA fragments showed no homology and presumably represented novel genes[33].
In summary, the findings of our study suggest that the lymphatic invasiveness of tumor cells is determined by multiple genes and co-factors with complex cellular signal pathways. Further functional study of the candidate novel genes might provide clues to molecular mechanism of tumor metastasis.
| 1. | Ji Y, Ling MY, Li Y, Xie H. Effect of cell fusion on metastatic ability of mouse hepatocarcinoma cell lines. World J Gastroenterol. 1999;5:22-24. [PubMed] |
| 2. | Diatchenko L, Lukyanov S, Lau YF, Siebert PD. Suppression subtractive hybridization: a versatile method for identifying differentially expressed genes. Methods Enzymol. 1999;303:349-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 243] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 3. | von Stein OD, Thies WG, Hofmann M. A high throughput screening for rarely transcribed differentially expressed genes. Nucleic Acids Res. 1997;25:2598-2602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 117] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 4. | Karlseder J. Telomere repeat binding factors: keeping the ends in check. Cancer Lett. 2003;194:189-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 58] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Deng Z, Atanasiu C, Burg JS, Broccoli D, Lieberman PM. Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein-Barr virus OriP. J Virol. 2003;77:11992-12001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 59] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 6. | Wu G, Jiang X, Lee WH, Chen PL. Assembly of functional ALT-associated promyelocytic leukemia bodies requires Nijmegen Breakage Syndrome 1. Cancer Res. 2003;63:2589-2595. [PubMed] |
| 7. | Iwano T, Tachibana M, Reth M, Shinkai Y. Importance of TRF1 for functional telomere structure. J Biol Chem. 2004;279:1442-1448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 83] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 8. | Kishi S, Lu KP. A critical role for Pin2/TRF1 in ATM-dependent regulation. Inhibition of Pin2/TRF1 function complements telomere shortening, radiosensitivity, and the G(2)/M checkpoint defect of ataxia-telangiectasia cells. J Biol Chem. 2002;277:7420-7429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 9. | Iizuka N, Mori N, Tamesa T, Tangoku A, Oka M. Telomerase activity and Nm23-H2 protein expression in hepatocellular carcinoma. Anticancer Res. 2003;23:43-47. [PubMed] |
| 10. | Nosaka K, Kawahara M, Masuda M, Satomi Y, Nishino H. Association of nucleoside diphosphate kinase nm23-H2 with human telomeres. Biochem Biophys Res Commun. 1998;243:342-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 11. | Li B, de Lange T. Rap1 affects the length and heterogeneity of human telomeres. Mol Biol Cell. 2003;14:5060-5068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 117] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 12. | Yang SW, Kim DH, Lee JJ, Chun YJ, Lee JH, Kim YJ, Chung IK, Kim WT. Expression of the telomeric repeat binding factor gene NgTRF1 is closely coordinated with the cell division program in tobacco BY-2 suspension culture cells. J Biol Chem. 2003;278:21395-21407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 13. | Yamada K, Yagihashi A, Yamada M, Asanuma K, Moriai R, Kobayashi D, Tsuji N, Watanabe N. Decreased gene expression for telomeric-repeat binding factors and TIN2 in malignant hematopoietic cells. Anticancer Res. 2002;22:1315-1320. [PubMed] |
| 14. | Otsuka T, Uchida N, Arima F, Shigematsu H, Fukuyama T, Maeda M, Sugio Y, Itoh Y, Niho Y. Down-regulation of human telomeric protein TRF1 gene expression during myeloid differentiation in human hematopoietic cells. Int J Hematol. 2000;71:334-339. [PubMed] |
| 15. | Fujimoto R, Kamata N, Taki M, Yokoyama K, Tomonari M, Nagayama M, Yasumoto S. Gene expression of telomerase related proteins in human normal oral and ectocervical epithelial cells. Oral Oncol. 2003;39:445-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 16. | Saito K, Yagihashi A, Nasu S, Izawa Y, Nakamura M, Kobayashi D, Tsuji N, Watanabe N. Gene expression for suppressors of telomerase activity (telomeric-repeat binding factors) in breast cancer. Jpn J Cancer Res. 2002;93:253-258. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 17. | Yamada M, Tsuji N, Nakamura M, Moriai R, Kobayashi D, Yagihashi A, Watanabe N. Down-regulation of TRF1, TRF2 and TIN2 genes is important to maintain telomeric DNA for gastric cancers. Anticancer Res. 2002;22:3303-3307. [PubMed] |
| 18. | Miyachi K, Fujita M, Tanaka N, Sasaki K, Sunagawa M. Correlation between telomerase activity and telomeric-repeat binding factors in gastric cancer. J Exp Clin Cancer Res. 2002;21:269-275. [PubMed] |
| 19. | Dokras A, Gardner LM, Kirschmann DA, Seftor EA, Hendrix MJ. The tumour suppressor gene maspin is differentially regulated in cytotrophoblasts during human placental development. Placenta. 2002;23:274-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 20. | Ngamkitidechakul C, Warejcka DJ, Burke JM, O'Brien WJ, Twining SS. Sufficiency of the reactive site loop of maspin for induction of cell-matrix adhesion and inhibition of cell invasion. Conversion of ovalbumin to a maspin-like molecule. J Biol Chem. 2003;278:31796-31806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 50] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 21. | Zhang M, Volpert O, Shi YH, Bouck N. Maspin is an angiogenesis inhibitor. Nat Med. 2000;6:196-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 323] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 22. | Biliran H Jr, Sheng S. Pleiotrophic inhibition of pericellular urokinase-type plasminogen activator system by endogenous tumor suppressive maspin. Cancer Res. 2001;61:8676-8682. [PubMed] |
| 23. | Zhang W, Zhang M. Tissue microarray analysis of maspin expression and its reverse correlation with mutant p53 in various tumors. Int J Oncol. 2002;20:1145-1150. [PubMed] |
| 24. | Khalkhali-Ellis Z, Hendrix MJ. Nitric oxide regulation of maspin expression in normal mammary epithelial and breast cancer cells. Am J Pathol. 2003;162:1411-1417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 25. | Maass N, Teffner M, Rösel F, Pawaresch R, Jonat W, Nagasaki K, Rudolph P. Decline in the expression of the serine proteinase inhibitor maspin is associated with tumour progression in ductal carcinomas of the breast. J Pathol. 2001;195:321-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 83] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 26. | Chen Z, Fan Z, McNeal JE, Nolley R, Caldwell MC, Mahadevappa M, Zhang Z, Warrington JA, Stamey TA. Hepsin and maspin are inversely expressed in laser capture microdissectioned prostate cancer. J Urol. 2003;169:1316-1319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 52] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Manzotti M, Dell'Orto P, Maisonneuve P, Zurrida S, Mazzarol G, Viale G. Reverse transcription-polymerase chain reaction assay for multiple mRNA markers in the detection of breast cancer metastases in sentinel lymph nodes. Int J Cancer. 2001;95:307-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 28. | Maass N, Hojo T, Rösel F, Ikeda T, Jonat W, Nagasaki K. Down regulation of the tumor suppressor gene maspin in breast carcinoma is associated with a higher risk of distant metastasis. Clin Biochem. 2001;34:303-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 80] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 29. | Sabbatini R, Federico M, Morselli M, Depenni R, Cagossi K, Luppi M, Torelli G, Silingardi V. Detection of circulating tumor cells by reverse transcriptase polymerase chain reaction of maspin in patients with breast cancer undergoing conventional-dose chemotherapy. J Clin Oncol. 2000;18:1914-1920. [PubMed] |
| 30. | Machtens S, Serth J, Bokemeyer C, Bathke W, Minssen A, Kollmannsberger C, Hartmann J, Knüchel R, Kondo M, Jonas U. Expression of the p53 and Maspin protein in primary prostate cancer: correlation with clinical features. Int J Cancer. 2001;95:337-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 31. | Yasumatsu R, Nakashima T, Hirakawa N, Kumamoto Y, Kuratomi Y, Tomita K, Komiyama S. Maspin expression in stage I and II oral tongue squamous cell carcinoma. Head Neck. 2001;23:962-966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 48] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 32. | Cui XN, Tang JW, Hou L, Song B, Li L, Liu JW. Screening differentially expressed genes in mouse hepatocarcinoma ascites cell line with high potential of lymphatic metastasis. World J Gastroenterol. 2005;11:1837-1842. [PubMed] |
| 33. | Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651-1656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1556] [Cited by in RCA: 1282] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
S- Editor Liu Y L- Editor Zhu LH E- Editor Ma WH