Published online Nov 7, 2006. doi: 10.3748/wjg.v12.i41.6702
Revised: July 12, 2006
Accepted: August 22, 2006
Published online: November 7, 2006
AIM: To study the Hepatitis B virus (HBV) genotypes and their effect on the progression and outcome in patients with chronic liver diseases from New Delhi, India.
METHODS: Sera from 100 HBV-related chronic liver disease (CLDB) cases were tested for HBV genotype using Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) and Type-specific primers-based PCR (TSP-PCR) targeting to the surface (S) gene encoding hepatitis B surface antigen.
RESULTS: Only genotypes A and D were present and genotype D was dominant. Genotype D was present in all CLDB patient categories. The genotype distribution for the 100 patients with CLDB was as follows: genotype A, 16/100 (16%) (7/40- 17% chronic hepatitis B (CHB); 8/47, 17%, HBV-related cirrhosis (CRB); 1/13, 7.6%, HBV-related hepatocellular carcinoma (HCCB); genotype D- 84/100 (84%) (32/40- 80% CHB; 38/47- 81%, CRB; 11/13, 85%, HCCB); genotype A + D, 3/100 (3%) (1/40- 3% CHB; 1/47- 2%, CRB; 1/13, 7.6%, HCCB); C, 0; B, 0; E, 0; F, 0; G 0, H 0; (P < 0.01, genotype D vs A).
CONCLUSION: Only HBV genotypes A and D were present in patients with CLDB from New Delhi, India. Compared with genotype D, genotype A patients had no significant clinical or biochemical differences (P > 0.05). Mixed infection with genotype A and D were seen in 3% of the cases. Genotype D was the dominant genotype prevalent in all patient categories.
- Citation: Chattopadhyay S, Das BC, Kar P. Hepatitis B virus genotypes in chronic liver disease patients from New Delhi, India. World J Gastroenterol 2006; 12(41): 6702-6706
- URL: https://www.wjgnet.com/1007-9327/full/v12/i41/6702.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i41.6702
Approximately, two billion people in the world have been infected by Hepatitis B virus (HBV), 350 million of whom are chronic carriers of the virus[1,2]. Worldwide HBV isolates have been classified into eight genotypes: A, B, C, D, E, F, G and H[1,3]. The eight genotypes have a characteristic geographical distribution[1]. Several studies have revealed the association of HBV genotypes with the severity of chronic liver disease, but the results are not consistent [1,4-6]. In Europe, most patients with genotype D are reported to have acute hepatitis B, while most patients with genotype A have chronic hepatitis B[5]. In northern Taiwan, 52% of asymptomatic carriers have genotype B, while 60% of patients with cirrhosis harbor genotype C; genotype B is more common in patients less than 50 years of age with HCC[7]. In Japan, HCC patients with genotype B younger than 60 years of age are rare[8]. Thus, the influences of HBV genotypes on clinical outcomes need to be clarified. Recent studies have shown that virological characteristics and clinical manifestations may differ even among HBV isolates of the same genotype[1]. Two subgroups of genotype B can be differentiated: a Japanese subgroup Bj with no indication for recombination events and an Asian subgroup called Ba which shows a recombination of the precore and core region from HBV genotype C. Response to antiviral therapies and the prevalence of hepatitis B e antigen (HBeAg) differ among patients with chronic liver diseases who are infected with HBV/Ba and HBV/Bj. Also, among isolates of HBV genotype A (HBV/A), two subtypes have been reported, one of which is distributed widely in European countries and the USA (Subtype A), while the other prevails in sub-Saharan Africa (Subtype A’). Subtype A’ seems to be virologically distinct from the original genotype A and is associated with reduced serum levels of HBV DNA and a low capacity to encode HBeAg. In addition, subtype A’ tends to induce hepatocellular carcinoma[1]. HBV genotype C isolates from Australian Aboriginees shows several features of a separate subgroup called genotype Caustralia. The South American genotype F segregates into two clades F1 and F2 [1].
Scanty information exists regarding the prevalence of HBV genotypes, their clinical relevance and therapeutic impact within CLDB category from India[9-12]. We have used recently developed two genotyping methods based on restriction fragment length polymorphism (RFLP)[13] and polymerase chain reaction (PCR) with type-specific primers[14] both targeting to the S-gene encoding the hepatitis B surface antigen.
The study subjects included HBV DNA positive patients belonging to different clinical categories: (1) The diagnosis of HBV-related cirrhosis (CRB) was made when the patients presented with features of HBV-related chronic liver disease associated with portal hypertension, had biochemical evidence of hepatocellular failure and barium swallow or endoscopy showed evidence of esophageal varices (When possible confirmation was done by histology). Other causes of portal hypertension were excluded on the basis of clinical features, liver function tests, and liver biopsy. The severity of the cirrhosis was quantified using the Childs-Pugh Classification strategy[15]. Forty-seven CRB cases were selected on the basis of the above-mentioned criteria. (2) Patients with infection of HBV showing symptomatic, biochemical (alanine aminotransferase more than upper limit of normal) or serological (hepatitis B surface Antigen (HBsAg), Hepatitis B e Antigen (HBeAg), IgG antibodies to hepatitis B core antigen (IgG- Anti- HBc) positivity), evidence of continued liver disease of more than 3 to 6 mo without steady improvement were diagnosed as suffering from Chronic Hepatitis B (CHB)[9-12,15]. Forty CHB cases were selected based on the above-mentioned criteria. (3) HBV related hepatocellular carcinoma (HCCB) was diagnosed in those HBV related cases when a percutaneous liver biopsy showed histological features of a primary malignancy of hepatocytes and/or alfa-fetoprotein levels of more than 30 nanogram/mL[1,12,15]. Thirteen HCCB cases were selected on the basis of the above-mentioned criteria.
With the assumption of prevalence of HBV, based on Indian Council of Medical Research (ICMR), Government of India, multicenter data[10-12], to be 70 % in chronic hepatitis (CH), 80% in liver cirrhosis (CRR), 60% in hepatocellular carcinoma (HCC), the minimum sample size required at 80% power level and 5% level of significance would be 30 cases each of HBV-related liver cirrhosis (CRB) and chronic hepatitis B (CHB) and 11 HBV-related hepatocellular carcinoma (HCCB). This sample size was achieved and some more cases were included by consecutive screening of 900 cases of various chronic liver disease patients over a period of 42 mo (June 2002 to December 2005). The study was approved by the institutional ethical committee of Maulana Azad Medical College, New Delhi. Study protocol was explained to all eligible participants and informed consent was obtained before subject enrollment.
Serological status was investigated by the third generation enzyme immuno-assay (EIA) method using the following commercially available EIA kits: hepatitis B surface antigen (HBsAg), hepatitis B e antigen (HBeAg), IgG antibodies to hepatitis B core antigen (IgG- Anti- HBc), IgG antibodies to hepatitis D (IgG anti-HDV) (Abbott Laboratories, Chicago, IL, USA) and anti-HCV by Innotest HCV AB III (Innogenetics NV, Ghent, Belgium).
Serum aspartate aminotransferase (AST), serum alanine aminotransferase (ALT), alkaline phosphatase (ALP), total bilirubin (T. Bil.), Prothrombin time (PT) tests were done weekly when the patients were admitted to the wards, while routine biochemical tests were done at monthly follow-up after the patients were discharged from the hospital.
In all CHB cases, Lamivudine monotherapy was provided. The drug was given at a dose of 100 mg per day for 18 mo. End of therapy response (ETR) in HBeAg positive patients was defined as loss of HBeAg (with or without the appearance of anti-HBe), reduction of viral DNA to undetectable level and normalization of ALT. However, in HBeAg negative/anti-HBe positive patients it was defined as loss of viral DNA accompanied with the normalization of ALT[16]. Sustained virological response (SVR) was considered when the status of ETR persisted for six months after the therapy was stopped [16].
Total nucleic acid from 100 μL serum was isolated using standard Proteinase-k/ phenol/ chloroform method[7]. Part of the S-gene was amplified by nested PCR as described earlier[13].
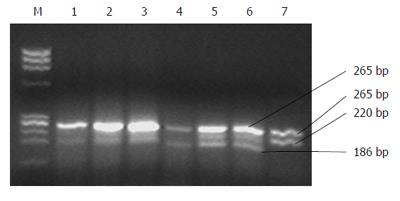
The second round PCR Product with a length of 485 bp was subjected to digestion with five kinds of restriction enzymes i.e. Ear I, Alw I, Nci I, Hph I, Nla IV[13].
Genotyping was done on the basis of PCR based type-specific DNA bands as performed earlier by Naito H et al 2001[14].
The amount of DNA corresponding to 100 μL of serum was dissolved in 25 μL of sterile deionised water and 10 μL was subjected to PCR. A 10-1 to 10-6 dilution of this sample was used to determine the sensitivity of the PCR protocol. Products were obtained up to 10-5 dilution. The detection limit of the PCR was calculated to be 200 HBV genomes/mL serum.
Representative PCR amplified products of S-gene (485 bases) were sequenced using the dideoxy chain termination method in an automated DNA sequencer (ABI 377-18 sequencing system, Perkin Elmer). Nucleotide sequences were aligned with CLUSTALW software. DNA sequences of Genotypes A-H were downloaded from the Gene bank according to the accession numbers for the respective genotype and phylogenetic analysis was carried out with the CLUSTALW algorithm.
Quantitative data was expressed as mean ± SD (Standard Deviation). Statistical significance was determined by Chi-square test with Yates’ correction (wherever needed) or by 2-sided Fisher exact test and Student’s t test using SPSS-10 software. The p values of less than 0.05 were considered significant throughout.
The S-gene of HBV isolates from 100 various CLDB patients were analyzed for HBV genotypes. The demographic and baseline clinical characteristics of the patients belonging to the three different groups (CHB, CRB, HCCB) is shown in Table 1. In all the three groups, the frequency of male patients was more than the female patients (Table 1). None of the patients was anti-HCV or anti-HDV positive as revealed by EIA tests.
| Parameters | CRB | CHB | HCCB |
| n | 47 | 40 | 13 |
| Male: Female | 41:6 | 36:4 | 11:2 |
| (6.8:1) | (9:1) | (5.5:1) | |
| Mean age (yr) | 38.36 ± 4.13a | 37.65 ± 9.49 | 47.80 ± 18.70 |
| ALT (nKat/ L) | 1333 ± 483 | 1200 ± 579.5 | 908.5 ± 363.4 |
| AST (nKat/L) | 1152 ± 620 | 1283.6 ± 483 | 971.4 ± 360 |
| Total Bil. (μmol/ L) | 0.00016 ± 0.00012 | 0.00147 ± 0.00218 | 0.0028 ± 0.00032 |
| IgG anti-HBc | 91% | 94% | 93% |
| Risk factors | |||
| Transfusions | 27% | 27% | 24% |
| Tattoo | 7% | 5% | 6% |
| Operation | 8% | 4% | 6% |
| Intravenous drug abuse | 5% | 6% | 3% |
| Unknown | 53% | 58% | 61% |
| Genotype A | (8/47) 17% | (7/40) 17% | (1/13) 7.6% |
| Genotype Db | (38/47) 81% | (32/40) 80% | (11/13) 85% |
| Genotype A+D | (1/47) 2% | (1/40) 3% | (1/13) 7.6% |
| HBeAg +ve (%) | 77%a | 82%a | 51% |
| Sustained virological response to therapy | Genotype A: (2/ 7) 28.5% | ||
| Genotype D: (12/32) 37.5% (P = NS) |
The PCR products of 100 HBV DNA positive subjects were digested with restriction enzymes specific for genotypes B, C, E and F and no characteristic fragment for each genotype was observed. Finally, by digestion with Nla IV, characteristic fragments of genotype A were observed in 16 subjects, characteristic fragments of genotype D were found in 81 subjects (Figure 1). Genotypes A+D mixed infections were found in three cases (Table 1).
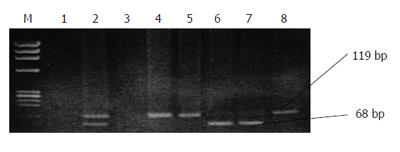
As explained in the methodology two sets of the 2nd PCR was carried out for detection of genotypes A to F. Sixteen (16%) of the 100 subjects had a PCR product of 68 bp that was characteristic of genotype A (Figure 2). The other 81/100 (81%) had PCR product a length of 119 bp and were classified into genotype D. Genotype A + D mixed infections were found in three cases (Table 1).
Twenty representative sequences were selected randomly from both genotype A (n = 5) and D (n = 15) samples. The 20 sequences were recorded in Gen Bank (Accession numbers: DQ875877 and DQ885267-DQ885285). A phylogenetic tree was constructed based on 410 bp of the S-gene of the HBV genome using CLUSTALW software. Out of the 20 isolates, 15 isolates were clustered with genotype D, and five were clustered with genotype A, validating both the genotyping strategies. The genotype A and D sequences were further classified into subtype A1 and Di by blast analysis. None of the sequences was clustered with the sequences of the genotypes B, C, E, F, G or H.
Sustained virological response on treatment with lamivudine was seen in 28.5% of patients of genotype A while it was seen in 37.5% of patients of genotype D (P value is not significant) (Table 1). The clinical data of CHB patients on therapy harboring either genotype A or genotype D did not differ significantly.
The majority of the cases turned out to be genotype D; 81/100 (81%), followed by genotype A; 16/100 (16%) using both the genotyping methods. Genotypes A + D mixed infections were found in three cases (Table 1). The ALT and AST levels, mean age, and HBeAg, IgG anti-HBc positivity were compared between genotypes A and D within each of the study groups and no significant differences were observed (Table 1). However, ALT, AST, HBeAg and IgG-anti-HBc positivity were significantly different between HCCB and CHB/ CRB cases (P < 0.05 CHB, CRB vs HCCB, Table 1).
There have been no studies from northern India comparing the clinical outcome of CLDB patients infected with different HBV genotypes using the TSP-PCR method. We have attempted to validate two most commonly used methods originally described by Naito et al and Mizokami et al[13,14]. Genotypes determined by both the methods were concordant in all the subjects.
The present study demonstrates that HBV genotype D is highly prevalent. This result, however, differs from that of two earlier studies on chronic liver diseases where genotype A and D were found to be prevalent in equal proportions[9,17]. The major reason of the different results might be due to the differential demographic distribution of the HBV-genotypes. The previous study was carried out in Lucknow; a city situated 600 km away from New Delhi, India. Also, the majority of the cases in Lucknow come from semi-urban and rural areas. The patients in New Delhi are more or less from urban or semi-urban areas[17]. Another reason could be selection bias, as earlier studies were conducted on small series of non-consecutive chronic cases [9,17].
In this study, mixed infection of genotypes A and D of HBV were found in 2% CRB cases, 3% CHB and 7.6% HCCB cases. This is in accordance with what has been reported earlier[17]. Interestingly, all the cases (3/3) with mixed infections with genotype A and D had histories of blood transfusions. In our study, genotype A was found only in the minority of the patient categories (Table 1). The impact of HBV genotypes on response to lamivudine therapy has been studied in various countries[18]. A study in Germany suggested that the rate of resistance to lamivudine was higher in patients with HBV genotype A infection than in patients with genotype D infection. No difference in the risk of lamivudine resistance is found between patients with genotype B and patients with genotype C. In patients with genotype C infection, however, virological response is worse during lamivudine therapy, and is also less durable after the discontinuation of therapy than in patients with genotype B infection[18]. The response to lamivudine is poorer in patients infected with subtype Ba, which contains a recombination with genotype C, than in those with subtype Bj without such a recombination[19]. Influence of genotypes on therapeutic response needs to be examined in patients infected with the other genotypes, particularly in those with genotype A or D infection[19]. In the present study, a significantly higher percentage of patients with CLDB were infected with genotype D and they did not influence the therapeutic response. Sustained virological response on treatment with lamivudine was seen in 28.5% of patients of genotype A while it was seen in 37.5% of patients of genotype D (P value is not significant). Determining the genotype could be helpful for predicting the outcome of antiviral therapy in patients with chronic hepatitis B. Transfusion was associated as the most important risk factor of HBV transmission in all the patient categories (Table 1).
Twenty representative samples from the study population were sequenced for the S-gene which led to confirmation of the RFLP and TSP-PCR results. Based on the nucleotide sequence of the S-region, genotype D emerged as the predominant genotype (15/20, 75%). Indian strains belonging to genotype A and D differed from each other by about 5.2% in the S-gene (410 bases).
In conclusion, genotype D appears to be the dominant genotype circulating in north Indian population with CLDB. Genotype A appears to be the minor genotype.
| 1. | Schaefer S. Hepatitis B virus: significance of genotypes. J Viral Hepat. 2005;12:111-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 167] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 2. | Kao JH. Hepatitis B viral genotypes: clinical relevance and molecular characteristics. J Gastroenterol Hepatol. 2002;17:643-650. [RCA] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 251] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 3. | Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575-2583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 725] [Cited by in RCA: 773] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 4. | Guettouche T, Hnatyszyn HJ. Chronic hepatitis B and viral genotype: the clinical significance of determining HBV genotypes. Antivir Ther. 2005;10:593-604. [PubMed] |
| 5. | Mayerat C, Mantegani A, Frei PC. Does hepatitis B virus (HBV) genotype influence the clinical outcome of HBV infection? J Viral Hepat. 1999;6:299-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 201] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 6. | Tsubota A, Arase Y, Ren F, Tanaka H, Ikeda K, Kumada H. Genotype may correlate with liver carcinogenesis and tumor characteristics in cirrhotic patients infected with hepatitis B virus subtype adw. J Med Virol. 2001;65:257-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 86] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 7. | Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology. 2000;118:554-559. [RCA] [DOI] [Full Text] [Cited by in Crossref: 684] [Cited by in RCA: 700] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 8. | Orito E, Ichida T, Sakugawa H, Sata M, Horiike N, Hino K, Okita K, Okanoue T, Iino S, Tanaka E. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology. 2001;34:590-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 315] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 9. | Thakur V, Guptan RC, Kazim SN, Malhotra V, Sarin SK. Profile, spectrum and significance of HBV genotypes in chronic liver disease patients in the Indian subcontinent. J Gastroenterol Hepatol. 2002;17:165-170. [RCA] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 216] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 10. | Tyagarajan SP, Jayaram S, Mohanavalli B. Prevalence of HBV in the general population of India. Hepatitis B in India. Problems and preventions. New Delhi, India: CBS 1996; 5-16. |
| 11. | Sarin SK, Chari S, Sundaram KR, Ahuja RK, Anand BS, Broor SL. Young v adult cirrhotics: a prospective, comparative analysis of the clinical profile, natural course and survival. Gut. 1988;29:101-107. [RCA] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 12. | Sundaram C, Reddy CR, Ramana GV, Benerjea S, Venkataratnam G, Kumari GS, Reddy BS, Bhaskaran CS. Hepatitis B surface antigen, hepatocellular carcinoma and cirrhosis in south India--an autopsy study. Indian J Pathol Microbiol. 1990;33:334-338. [PubMed] |
| 13. | Mizokami M, Nakano T, Orito E, Tanaka Y, Sakugawa H, Mukaide M, Robertson BH. Hepatitis B virus genotype assignment using restriction fragment length polymorphism patterns. FEBS Lett. 1999;450:66-71. [RCA] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 184] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 14. | Naito H, Hayashi S, Abe K. Rapid and specific genotyping system for hepatitis B virus corresponding to six major genotypes by PCR using type-specific primers. J Clin Microbiol. 2001;39:362-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 211] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 15. | Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, Denk H, Desmet V, Korb G, MacSween RN. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696-699. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3521] [Cited by in RCA: 3838] [Article Influence: 123.8] [Reference Citation Analysis (2)] |
| 16. | Lok AS, McMahon BJ. Chronic hepatitis B. Hepatology. 2001;34:1225-1241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 642] [Cited by in RCA: 639] [Article Influence: 25.6] [Reference Citation Analysis (1)] |
| 17. | Kumar A, Kumar SI, Pandey R, Naik S, Aggarwal R. Hepatitis B virus genotype A is more often associated with severe liver disease in northern India than is genotype D. Indian J Gastroenterol. 2005;24:19-22. [PubMed] |
| 18. | Enomoto M, Tamori A, Nishiguchi S. Hepatitis B virus genotypes and response to antiviral therapy. Clin Lab. 2006;52:43-47. [PubMed] |
| 19. | Akuta N, Kumada H. Influence of hepatitis B virus genotypes on the response to antiviral therapies. J Antimicrob Chemother. 2005;55:139-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
S- Editor Pan BR L- Editor Ma JY E- Editor Ma WH