Published online Nov 7, 2006. doi: 10.3748/wjg.v12.i41.6614
Revised: August 12, 2006
Accepted: October 6, 2006
Published online: November 7, 2006
AIM: To establish a multidrug resistant (MDR) cell sub-line from the human hepatocarcinoma cell line (HepG2) in nude mice.
METHODS: HepG2 cell cultures were incubated with increasing concentrations of adriamycin (ADM) to develop an ADM-resistant cell subline (HepG2/ADM) with cross-resistance to other chemotherapeutic agents. Twenty male athymic BALB/c-nu/nu mice were randomized into HepG2/nude and HepG2/ADM/nude groups (10 in each group). A cell suspension (either HepG2 or HepG2/ADM) was injected subcutaneously into mice in each group. Tumor growth was recorded, and animals were sacrificed 4-5 wk after cell implantation. Tumors were prepared for histology, and viable tumor was dispersed into a single-cell suspension. The IC50 values for a number of chemotherapeutic agents were determined by 2, 3-bis (2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide inner salt (MTT) assay. Rhodamine-123 retention/efflux and the level of resistance-associated proteins were determined by flow cytometry. The mRNA expression of mdr1, mrp and lrp genes was detected using reverse transcriptase polymerase chain reaction (RT-PCR) in HepG2/nude and HepG2/ADM/nude groups.
RESULTS: The appearances of HepG2/nude cells were slightly different from those of HepG2/ADM/nude cells. Similar tumor growth curves were determined in both groups. A cross-resistance to ADM, vincristine, cisplatin and 5-fluorouracil was seen in HepG2/ADM/nude group. The levels of P-glycoprotein and multidrug resistance-associated proteins were significantly increased. The mRNA expression levels of mdr1, mrp and lrp were higher in HepG2/ADM/nude cells.
CONCLUSION: ADM-resistant HepG2 subline in nude mice has a cross resistance to chemotherapeutic drugs. It may be used as an in vivo model to investigate the mechanisms of MDR, and explore the targeted approaches to overcoming MDR.
- Citation: Zhai BJ, Shao ZY, Zhao CL, Hu K, Wu F. Development and characterization of multidrug resistant human hepatocarcinoma cell line in nude mice. World J Gastroenterol 2006; 12(41): 6614-6619
- URL: https://www.wjgnet.com/1007-9327/full/v12/i41/6614.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i41.6614
Tumor cell drug resistance represents a significant obstacle to successful chemotherapy. Cells which have acquired resistance to one anti-tumor drug usually show resistance to other anti-tumor drugs[1]. This cellular resistance is known as multidrug resistance (MDR). Overexpression of P-glycoprotein (P-gp), multidrug resistance-associated protein (MRP), and lung resistance-related protein (LRP) is associated with development of MDR in cancer cells[2]. P-gp acts as an energy-dependent outward transport pump and can decrease intracellular drug accumulation by removing chemotherapeutic drugs from the cytoplasm[3]. A correlation between mdr1 mRNA expression and drug resistance has been demonstrated using human cancer cell lines[4]. MRP is an ATP-dependent transmembrane protein related to MDR[5], and LRP is also expressed in a MDR lung cancer line[6]. The mrp and lrp genes encoding MRP and LRP, respectively, have been cloned in MDR cell lines that do not exhibit P-gp overexpression.
Several cell lines from resistant carcinomas have been established to elucidate the mechanisms of MDR. MDR cell lines and the expression of three resistance-associated markers (P-gp, MRP and LRP) have been extensively studied in vitro[7]. These resistant tumor cell lines have been established in vitro from P-gp negative cell lines via the exposure of chemotherapeutic drugs such as adriamycin (ADM), and their acquired resistance is mainly dependent on P-gp expression levels[8]. Unfortunately, it seems that the predictive value of MDR in established cell lines in vitro for determining the responsiveness of a relative tumor in clinical practice is poor. This misinterpreted role of MDR may explain the failure of in vivo strategies to reverse MDR by using modulators in solid tumors[9,10]. The aim of this study was to establish a multidrug resistant subline from a human hepatocellular carcinoma line (HepG2) in nude mice, and to acquire more insights into in vivo drug resistance.
2, 3-bis (2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide inner salt (MTT), ADM, vincristine (VCR), cisplatin, (CDDP), 5-fluorouracil (5-Fu), 2-(6-amino-3-imino-3H-xanthen-9-yl)-benzoic acid methyl ester (rhodamine123, Rh123), and dimethyl sulphoxide (DMSO) were obtained from Sigma (St Louis, Missouri, USA). Mouse monoclonal antibody MRK16 was supplied by Santa Cruz Biotechnology (Santa Cruz, California, USA). Rat monoclonal antibody MRPr1 and mouse monoclonal antibody LRP56 were obtained from Caltag Laboratories (Burlingame, California, USA). Fluorescein isothiocynate (FITC)-labeled goat anti-mouse immunoglobulin G (IgG) was from Tago Immunologicals (Camarillo, California, USA), and mouse monoclonal IgG was from Chemicon (Temecula, California, USA). TRIzol, RT-PCR kit, and primers were supplied by Life Technologies Inc. (Rockville, Minnesota, USA).
HepG2 was provided by the Institute of Biochemistry and Cell Biology (Chinese Academy of Sciences, Shanghai, China). Cells were grown as monolayers in RPMI 1640 medium, supplemented with 10% (v/v) heat-inactivated fetal calf serum, 1.0 mmol/L sodium pyruvate at 37°C in a humidified atmosphere containing 50 mL CO2. ADM was added to HepG2 cell cultures in stepwise increasing concentrations (starting at 0.001 mg/L and ending up to 1.0 mg/L) to develop a drug resistant cell subline (HepG2/ADM). After removal of dead cells, the remaining viable cells were identified as being drug resistant, and then cultured in a higher concentration of anticancer drug again. With the gradual increase in drug concentration, cells could finally be maintained in a culture medium containing 1000 μg/L ADM. The MDR characteristics of these HepG2/ADM cells were tested at various concentrations of anticancer drugs including ADM, VCR, CDDP and 5-Fu.
The animal study was approved by the Experimental Animal Committee of Chongqing University of Medical Sciences, and all animal experiments adhered to the Animal Welfare Committee guidelines. Male athymic BALB/c nu/nu mice (4-6 wk old) were obtained from the Institute of Materia Medica (Chinese Academy of Sciences, Shanghai, China) and housed in laminar-flow cabinets under specific pathogen-free (SPF) conditions.
Twenty male athymic BALB/c-nu/nu mice were randomized into HepG2/nude and HepG2/ADM/nude groups, 10 in each group. A suspension of either HepG2 or HepG2/ADM cells (107 cells in 0.15 mL of Hanks’ solution) was injected into the right subaxillary region of mice in each group. After implantation, the tumor growth was detected daily by measuring its diameter with Vernier caliper. Tumor weight (TW) was calculated using the following formula: TW (mg) = tumor volume (mm3) = d2 × D/2, where d and D are the shortest and longest diameters, respectively. Animals were sacrificed 4-5 wk after implantation when the subcutaneous tumor reached the size of 1.5 cm in diameter, and samples were harvested. Tumor tissue blocks were created for histological examinations. Viable tumor tissue was ground into a single-cell suspension in a loose-fitting ground glass homogenizer. Cell suspensions were washed twice with DMEM, and cultured in RPMI 1640 medium supplemented with 10% fetal calf serum.
MTT cell proliferation assay was performed to determine the percentage of viable HepG2 or HepG2/ADM cells in vitro, or HepG2/nude and HepG2/ADM/nude cells were derived from viable tumor tissues in vivo after incubation with various concentrations of anti-tumor drugs. In brief, cell suspensions were seeded in 96-well culture plates at a density of 1 × 105 cells/well. After 24 h incubation, the cells were exposed to the drug under study for 72 h, followed by 4 h incubation with the tetrazolium staining. Using a multiwell spectrophotometer reader (Molecular Devices, Menlo Park, California, USA), the optical intensity at 570 nm was measured, and cell viability was assessed in each well from the level of a dark blue formazan crystal created. Each assay was performed in triplicate, and RPMI 1640 medium was used as a blank control. The IC50 values were defined as the concentration resulting in 50% cell survival. After the dose-response curve was plotted, the IC50 for each anti-tumor drug was determined. The relative resistance to anti-tumor drugs was determined by dividing the IC50 values obtained for HepG2/ADM cells by those of HepG2 cells.
A total of 1 × 106 HepG2 or HepG2/ADM cells were seeded into each well of a 12-well culture plate, and incubated for 24 h. After the culture medium was replaced by Hanks' solution supplemented with 10 mmol/L HEPES buffer and 10% fetal calf serum (FCS), the cells were incubated for 15 min at 37°C. Rhodamine 123 (Rh123) was then added to give a final concentration of 10 g/L. The cells were further incubated for 30 min, washed twice with ice-cold PBS, harvested with trypsin, and suspended in PBS. The cells were further incubated in Rh123-free medium for 30 min to determine the Rh123 efflux. Cell surface-associated fluorescence was measured after addition of ice-cold Rh123 to the cells. Using the FACS caliver system (Elite ESP, Coulter Electronics, Miami, USA), the fluorescence intensity of Rh123 was measured at excitation and emission wavelength of 485 nm and 530 nm, respectively.
The protein levels of P-gp, MRP and LRP were determined by flow cytometry using the monoclonal antibodies MRK16, MRPr1 and LRP-56. A total of 1 × 106 HepG2 or HepG2/ADM cells were incubated in suspension with 10 g/L of the primary antibodies and an isotype-matched mouse IgG2a, respectively, for 1 h at 37°C. After washing, the cells were incubated with 0.175 g/L FITC-labeled goat anti-mouse IgG (for P-gp and LRP-56) or FITC-labeled goat anti-rat IgG (for MRPr1) for 1 h at 37°C. The fluorescence intensity (excitation wavelength 488 nm) was determined using an Epics ESP flow cytometer (Coulter Electronics, Miami, USA). The ratio of the specific fluorescence intensity of the HepG2/ADM cells to that of the HepG2 cells indicated the relative level of resistance-associated proteins.
Reverse transcriptase polymerase chain reaction (RT-PCR) was used to measure the mRNA expression of mdr1, mrp and lrp genes. The primers of mdr1, mrp and lrp are as follows: mdr1: 5’-GGC TCC GAT ACA TGG TTT TCC-3’, 3’-TTC AGT GCG ATC TTC CCA GC-5’; mrp: 5’-TGA AGG ACT TCG TGT CAG CC-3’, 5’ GTC CAT GAT GGT GTT GAG CC-3’; lrp: 5’-CCT CGA GAT CCA TTG TGC TGG-3’, 5’-CAC AGG GTT GGC CAC TGT GCA-3’. β-actin expression was used as a control for the amount of RNA used, and its primer was 5’-ACC CCC ACT GAA AAA GAT GA-3’, and 5’-ATC TTC AAA CCT CCA TGA TG-3’. Total RNA was extracted from cells using TRIzol. mdr1, mrp, lrp and β-actin RNA transcripts were detected using RT-PCR. An aliquot of each reaction mixture was then analyzed by electrophoresis using 1.5% agarose gel. Densitometry was performed with an UVP gel image analysis system (BIO-RAD, Melville, NY, USA) and the ratio between the target and control PCR products was determined by dividing the densitometric volume of the target band by that of the control band.
The PCR cycle included heat denaturation at 94°C for 45 s, annealing at 60°C for 45 s, and polymerization at 72°C for 90 s. cDNA was prepared from 4.5 mg of total RNA using the GeneAmp RNA PCR kit. The reaction mixture was incubated at 42°C for 30 min and then heated for 5 min at 99°C to inactivate MMLV reverse transcriptase. The cDNA derived from 0.15 mg of total RNA was mixed with 20 mL reaction mixture (10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl, 1.25 mmol/L MgCl2, 188 nmol/L gene-specific primers and 0.625 U AmpliTaq DNA polymerase) with β-actin (0.765 amol). Thirty-two cycles of PCR were performed, each consisting of heat denaturation at 94°C for 45 s, annealing at 55°C for 1 min, and polymerization at 72°C for 2 min.
All observed data were expressed as mean ± SD. The statistical significance of any observed difference between the mean values of HepG2 and HepG2/ADM groups was evaluated using the Student’s t test. P < 0.05 was considered statistically significant.
Both HepG2/nude and HepG2/ADM/nude tumor cells demonstrated polygonal epithelial-like morphology, with firm attachment to the culture flask. They had conspicuous large nuclei. Electron microscopy showed abundant microvilli and projections on the cell surface in both cell lines. Some of the projections on HepG2/nude cells were long, terminated with swellings, while those of HepG2/ADM/nude cells were short and compact. Both cell types had many lysosomes in the cytoplasm and were usually concentrated on one side of the HepG2/nude cells, but scattered around the cytoplasm in HepG2/ADM/nude cells. No obvious desmosomes and tight junctions were observed in the cells.
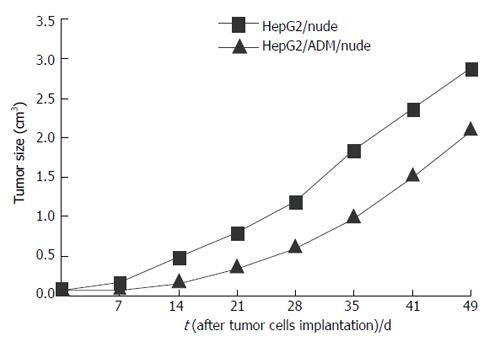
Tumor growth was evaluated in nude mice after subcutaneous implantation of HepG2/nude and HepG2/ADM/nude cells. Mean tumor volumes measured with a Vernier caliper are shown in Figure 1.
HepG2/ADM tumor cells became drug resistant following incubation of HepG2 cells with increasing concentrations of ADM (Table 1). In addition to direct resistance to ADM, these cells were also resistant to other chemotherapeutic agents including VCR, 5-FU and CDDP. The IC50 values for the drugs were significantly greater in the HepG2/ADM group than in the HepG2 group (Table 1), indicating that HepG2/ADM tumors awarded MDR characteristics to nude mice. However, the resistant index for ADM and VCR was significantly lower in the HepG2/ADM/nude group than in the HepG2/ADM group (ADM: 26.31 vs 51; VCR: 6.17 vs 57.06), but that for CDDP and 5-Fu was higher (CDDP: 6.45 vs 2.33; 5-Fu: 4.09 vs 1.49).
| Drug | IC50 values (mg/L) | RIa | RIb | |||
| HepG2 | HepG2/ADM | HepG2/nude | HepG2/ADM/nude | |||
| ADM | 0.02 ± 0.015 | 1.02 ± 0.326b | 0.16 ± 0.015 | 4.21 ± 0.012d | 51.00 | 26.31 |
| VCR | 0.18 ± 0.011 | 10.27 ± 2.415b | 0.18 ± 0.211 | 1.11 ± 0.075d | 57.06 | 6.17 |
| CDDP | 0.87 ± 0.023 | 2.03 ± 0.360 | 0.22 ± 0.023 | 1.42 ± 0.032d | 2.33 | 6.45 |
| 5-FU | 9.48 ± 0.112 | 14.16 ± 1.924 | 0.53 ± 0.112 | 2.17 ± 0.670 | 1.49 | 4.09 |
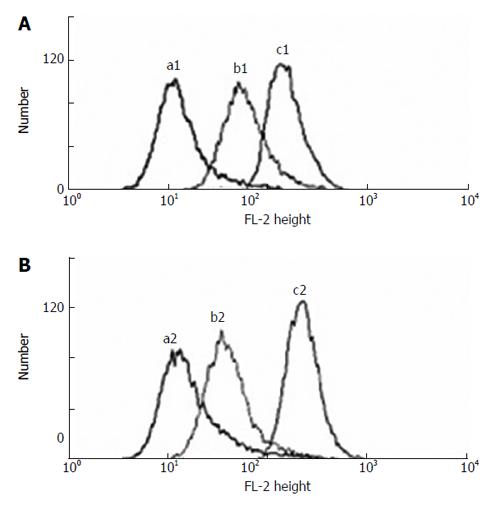
Compared to both HepG2 and HepG2/nude tumor cells, either decreased retention or increased efflux of Rh123 was observed in the HepG2/ADM and HepG2/ADM/nude tumor cells in nude mice. The pattern of altered retention or efflux of Rh123 in the ADM resistant cells is shown in Figure 2.
The relative expressions of resistance-associated proteins were given as the ratios of the specific fluorescence intensity of HepG2/ADM and HepG2/ADM/nude cells to those of HepG2 and Hep2/nude cells respectively. As shown in Table 2, HepG2/ADM/nude tumors co-expressed the resistance-associated proteins, P-gp, MRP and LRP respectively. Compared to the HepG2/nude cells, the levels of P-gp and MRP were significantly increased in the HepG2/ADM/nude cells (P < 0.01), whereas no significant change in LRP expression was observed between the HepG2/nude and HepG2/ADM/nude groups (P > 0.05).
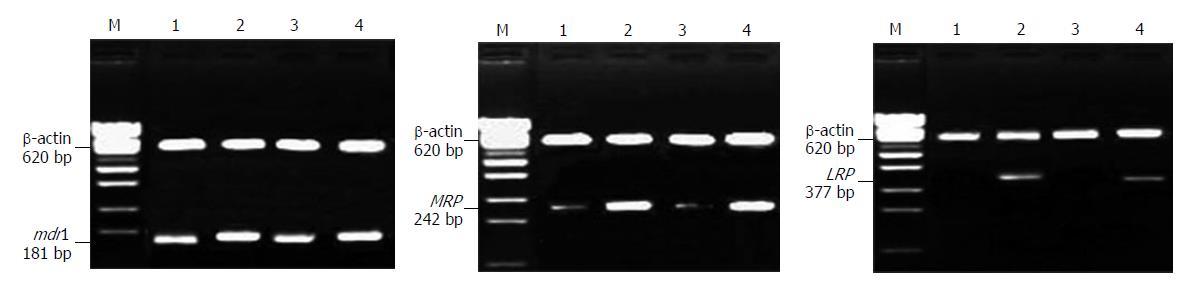
Positive mRNA expression of the mdr1 and mrp genes and negative mRNA expression of the lrp gene were seen in both HepG2 and HepG2/nude cells. However, mRNA expression of the mdr1, mrp and lrp genes was greater in HepG2/ADM and HepG2/ADM/nude tumor cells than in HepG2 and HepG2/nude tumor cells (Figure 3).
As drug resistance is one of the major barriers to the successful treatment of malignancies, investigation of the mechanisms of drug resistance and approaches to overcoming it has been widely performed in the past decades. Many MDR cell lines have been established in vitro as clinically relevant cancer models in these studies. However, compared to in vitro studies, the results in vivo are unsatisfactory in the treatment of solid tumors with modulators of Pg-p[9]. It appears that the predictive value of MDR in vitro is not so good for determining the responsiveness of tumors in clinical practice[10]. In addition to tumor heterogeneity in the expression of MDR[11], several reasons are given for these negative results. The interface between MDR modulators and chemotherapeutic agents increases drug toxicity[12]. Furthermore, MDR represents the net effect of the expression of a variety of genes involved in the development of drug resistance[13,14]. Finally, some MDR results obtained in ex vivo studies have been overvalued or misunderstood[15]. Tumor cells grown in solid tumors in vivo are less vulnerable to drugs than the same cells grown in monolayer culture in vitro due to inadequate drug penetration, a reduced growth fraction and a decreased sensitivity mediated by cell-cell interactions. It is therefore important to establish a well-characterized and standardized MDR model in vivo for drug screening and prediction of clinical drug response.
In this study, we established an ADM-resistant subline in vitro from the human HepG2 cell line with increasing concentrations of ADM. This subline (HepG2/ADM) demonstrated cross resistance to other chemotherapeutic drugs. The MTT assay showed that HepG2/ADM cells not only presented direct resistance to MDR-related drugs such as ADM (51-fold) and VCR (57-fold), but also were sensitive to non-MDR-related compounds such as CDDP (2.33-fold) and 5-Fu (1.49-fold). Furthermore, it showed characteristics of MDR tumor cells, including expression of MDR-associated genes, and presence of resistance-associated proteins. However, it is important to confirm whether these resistant characteristics could be maintained in vivo following their implantation in nude mice.
Slight differences were observed under light micro-scope in HepG2/nude cells, and HepG2/ADM/nude tumor cells. These changes became more apparent under electron microscope, especially in the shape of cellular microvilli and the distribution of lysosomes in the cytoplasm. These changes had no significant effects on tumor growth between the two experimental groups. However, the in vivo tumor growth rates were almost similar between the two groups. Furthermore, the chemo-resistance in vitro was confirmed in vivo after subcutaneous implantation of HepG2/ADM cells in nude mice. The IC50 values for ADM and VCR were significantly higher in the HepG2/ADM group than in the HepG2/ADM/nude group, but those for CDDP and 5-Fu were lower in the HepG2/ADM group than in the HepG2/ADM/nude group, indicating that a partial transfer of resistance ability acquired ex vivo to the in vivo situation is possible.
In the present study, a number of proteins involved in the development of drug resistance were analyzed quantitively using flow cytometry to determine the level of MDR-associated proteins in vivo, including P-gP, MRP and LRP. The levels of P-gP, MRP and LRP were significantly higher in the HepG2/ADM/nude subline than in the HepG2/nude cells, but no statistical difference was seen between HepG2/ADM and HepG2/ADM/nude groups. Similar results were seen in the in vivo expression of MDR-associated genes (mdr1, mrp and lrp) on mRNA levels. Although RT-PCR used in this study was semi-quantitative, it showed the mRNA expression in HepG2/ADM/nude tumor cells induced by ADM. These findings confirmed quantitively by both immunoflow cytometry might be beneficial for protein and functional rhodamine assays.
However, it still remains unclear why the resistant index changed significantly in ex vivo conditions compared to in vivo conditions. This can be probably explained by the assumption that other MDR mechanisms are involved in the subline[16], such as the increase of ATP dependent glutathione S-conjugate transporter[17], and the decrease of DNA topoisomerase II activity[14].
In conclusion, a MDR model of nude mice can be successfully established using human hepatocarcinoma cell line HepG2. This new model shows resistance to chemotherapeutic drugs, and mRNA expression of various MDR-associated genes on the protein levels. It can be used as an in vivo model to investigate the molecular mechanisms involved in MDR-related genes of hepatocarcinoma and to explore the targeted approaches tor overcoming MDR in tumor cells.
| 1. | Bradley G, Ling V. P-glycoprotein, multidrug resistance and tumor progression. Cancer Metastasis Rev. 1994;13:223-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 159] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 2. | van Brussel JP, van Steenbrugge GJ, Romijn JC, Schröder FH, Mickisch GH. Chemosensitivity of prostate cancer cell lines and expression of multidrug resistance-related proteins. Eur J Cancer. 1999;35:664-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 82] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 3. | Takara K, Sakaeda T, Okumura K. An update on overcoming MDR1-mediated multidrug resistance in cancer chemotherapy. Curr Pharm Des. 2006;12:273-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 175] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 4. | Monks A, Scudiero D, Skehan P, Shoemaker R, Paull K, Vistica D, Hose C, Langley J, Cronise P, Vaigro-Wolff A. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J Natl Cancer Inst. 1991;83:757-766. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2404] [Cited by in RCA: 2447] [Article Influence: 69.9] [Reference Citation Analysis (0)] |
| 5. | Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, Stewart AJ, Kurz EU, Duncan AM, Deeley RG. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258:1650-1654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2152] [Cited by in RCA: 2217] [Article Influence: 67.2] [Reference Citation Analysis (0)] |
| 6. | Scheffer GL, Wijngaard PL, Flens MJ, Izquierdo MA, Slovak ML, Pinedo HM, Meijer CJ, Clevers HC, Scheper RJ. The drug resistance-related protein LRP is the human major vault protein. Nat Med. 1995;1:578-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 423] [Cited by in RCA: 430] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 7. | Izquierdo MA, Shoemaker RH, Flens MJ, Scheffer GL, Wu L, Prather TR, Scheper RJ. Overlapping phenotypes of multidrug resistance among panels of human cancer-cell lines. Int J Cancer. 1996;65:230-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 8. | Hill BT. Differing patterns of cross-resistance resulting from exposures to specific antitumour drugs or to radiation in vitro. Cytotechnology. 1993;12:265-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 9. | Pinedo HM, Giaccone G. P-glycoprotein--a marker of cancer-cell behavior. N Engl J Med. 1995;333:1417-1419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 56] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 10. | Thomas H, Coley HM. Overcoming multidrug resistance in cancer: an update on the clinical strategy of inhibiting p-glycoprotein. Cancer Control. 2003;10:159-165. [PubMed] |
| 11. | Di Nicolantonio F, Mercer SJ, Knight LA, Gabriel FG, Whitehouse PA, Sharma S, Fernando A, Glaysher S, Di Palma S, Johnson P. Cancer cell adaptation to chemotherapy. BMC Cancer. 2005;5:78. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 108] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 12. | Krishna R, Mayer LD. Modulation of P-glycoprotein (PGP) mediated multidrug resistance (MDR) using chemosensitizers: recent advances in the design of selective MDR modulators. Curr Med Chem Anticancer Agents. 2001;1:163-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 13. | Muller M, Meijer C, Zaman GJ, Borst P, Scheper RJ, Mulder NH, de Vries EG, Jansen PL. Overexpression of the gene encoding the multidrug resistance-associated protein results in increased ATP-dependent glutathione S-conjugate transport. Proc Natl Acad Sci USA. 1994;91:13033-13037. [RCA] [DOI] [Full Text] [Cited by in Crossref: 388] [Cited by in RCA: 419] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 14. | Volm M, Kastel M, Mattern J, Efferth T. Expression of resistance factors (P-glycoprotein, glutathione S-transferase-pi, and topoisomerase II) and their interrelationship to proto-oncogene products in renal cell carcinomas. Cancer. 1993;71:3981-3987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 15. | Kolfschoten GM, Hulscher TM, Pinedo HM, Boven E. Drug resistance features and S-phase fraction as possible determinants for drug response in a panel of human ovarian cancer xenografts. Br J Cancer. 2000;83:921-927. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 16. | Uchiyama-Kokubu N, Watanabe T. Establishment and characterization of adriamycin-resistant human colorectal adenocarcinoma HCT-15 cell lines with multidrug resistance. Anticancer Drugs. 2001;12:769-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 17. | Paul S, Breuninger LM, Tew KD, Shen H, Kruh GD. ATP-dependent uptake of natural product cytotoxic drugs by membrane vesicles establishes MRP as a broad specificity transporter. Proc Natl Acad Sci USA. 1996;93:6929-6934. [RCA] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
S- Editor Wang GP L- Editor Wang XL E- Editor Bi L