INTRODUCTION
Hepatitis B virus (HBV) infection is a major cause of liver disease worldwide, ranging from acute and chronic hepatitis to cirrhosis and hepatocellular carcinoma[1]. HBV replication involves reverse-transcription of a pregenomic RNA (pgRNA) intermediate inside nucleocapsids, which are formed by 180 or 240 core protein subunits. Inside the capsid, the viral polymerase converts pgRNA into minus-strand DNA, which in turn is completed to a double-stranded, relaxed circular DNA molecule[2,3]. This life cycle places HBV into the large family of retroelements, which all require reverse-transcription of an RNA intermediate.
Recently, a cellular defense mechanism targeting a wide range of retroviruses was identified. It was shown that the propagation of HIV-1 strains lacking the accessory protein Vif is suppressed in nonpermissive cells due to expression of the cytidine deaminase APOBEC3G[4,5]. Further studies revealed that A3G induces massive C→U deamination of single stranded retroviral DNA, resulting in DNA degradation or lethal G→A hypermutation[6-8]. In addition, APOBEC3F (another cytidine deaminase of APOBEC family) is also a potent retroviral restrictor, but its activity, unlike that of APOBEC3G, is partially resistant to HIV-1 Vif and results in retroviral hypermutation. Moreover, APOBEC3F and APOBEC3G appear to be coordinately expressed in a wide range of human tissues and are independently able to inhibit retroviral infection. Thus, APOBEC3F and APOBEC3G are likely to function alongside one another in the provision of an innate immune defense, with APOBEC3F functioning as the major contributor to HIV-1 hypermutation in vivo[9]. Interestingly, APOBEC3G can also interfere with the HBV life cycle in cotransfected cells[10,11]. Surprisingly, however, reduced levels of encapsidated viral pgRNA rather than extensive editing were found to be the major contributing factor[10]. APOBEC3G-mediated editing did occur but was only detected in a minority of clones produced in transfected HepG2 hepatoma cells[11].
In the present study, we performed a detailed analysis of the inhibition effect of APOBEC3G on hepatitis B virus replication in cell culture and in an HBV vector-based mouse model.
MATERIALS AND METHODS
Plasmid constructs
To construct expression vectors coding for human APOBEC3G (pXFA3G), total RNAs were extracted from peripheral blood mononuclear cells (PBMCs). RT-PCR amplification of human APOBEC3G sequence used forward primer 5’-CGGAATTCAAGCCTCACTTCAGAAACAC-3’ and reverse primer 5’- CGAAGCTTTCTGCCTTCCTTAGAGACT G-3’. The PCR product was cloned into EcoRI/HindIII restriction sites of the CMV-driven expression vector fused with a hemagglutinin fusion epitope tag at its carboxy terminal end (pXF3H). Replication competent wild-type HBV 1.3 fold over-length plasmid (subtype, ayw) has been constructed previously in our laboratory.
Cell culture, transfection and harvesting
HuH-7 cells were grown at 37°C with 50 mL/L CO2 in Dulbecco’s modified Eagle medium, supplemented with 10% fetal calf serum (Invitrogen, CA, USA). The cells were plated at a density of 4.5 × 105 cells per well in 6-well plates 18 h prior to transfection. Transfecion of cells was performed with lipofectamine 2000 (Invitrogen, CA, USA) following the user guidelines. On d 3 after transfection, the cells were removed from the culture dish by treatment with trypsin-EDTA, resuspended in culture medium, washed with phosphate-buffered saline, pelleted, and resuspended in 1 mL chilled lysis buffer (140 mmol/L NaCl, 1.5 mmol/L MgCl2, 50 mmol/L Tris-HCl [pH 8.0]) containing 0.5% Nonidet P-40. Nuclei were removed via centrifugation for 5 min at 2000 r/min in an Eppendorf centrifuge, and the supernatant was cleared of cell debris by centrifugation for another 5 min at 14 000 r/min
HBV vector-based mouse model
For the in vivo experiments, 6 to 8 wk-old female BALB/c mice were used. A total of 18 mice were randomly divided into 3 groups(6 mice in each). Replication competent pHBV1.3 (10 μg) and APOBEC3G expression vectors pXFA3G (10 μg) or pXF3H (10 μg) control plasmid DNA were co-injected into the tail vein of mice in a volume of Ringer’s injection equivalent to 9% of the mouse body weight[12,13]; the total volume was delivered within 5 s. The mouse sera were collected at indicated time after hydrodynamic injection, and secreted HBsAg levels and HBV DNA content were measured. All mouse experiments were carried out according to the guidelines established by the Institutional Animal Care and Use Committee at the Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China.
HBsAg and HBeAg assays and western blot analysis
Levels of HBsAg and HBeAg in the media of the transfected cells, and in the sera (1:100 diluted) of the treated mice were determined using ELISA (Shanghai Shiye Kehua Company, China). For western blot analysis, cytoplasmic lysates were incubated with 1 volume 2 x loading buffer containing 10% beta-mercaptoethanol for 10 min at 95°C before loading onto a 12.5% SDS-PAGE. Proteins were transferred onto nitrocellulose membrane via electroblotting. The membranes were incubated with HBV core specific rabbit antiserum (Santa Cluz,USA) or with anti-hemagglutinin fusion epitope monoclonal antibody (Santa Cluz, USA) followed by horseradish peroxidase-conjugated mouse anti-rabbit antibody. Proteins were visualized via enhanced chemiluminescence (Roche, Germany).
HBV DNA purification and analysis
The method for purification of cytoplasmic core-associated HBV DNA was adapted from Pugh et al[14]. Briefly, Huh7 cells were disrupted in lysis buffer (100 mmol/L Tris- HCl pH 8.0, 0.2% NP-40). The cell lysate was clarified by centrifugation at 13 000 r/min for 1 min to pellet nuclei and insoluble material. The supernatant was adjusted to 6 mmol/L MgOAc2 and incubated for 2 h at 37°C with 200 μg/mL of DNase I and 100 μg/mL of RNase A. Following digestion, the lysate was centrifuged for 1 min at 13 000 r/min. The supernatant was incubated for 1 h at 55°C after addition of 10 mmol/L EDTA, 1% SDS, 100 mmol/L NaCl and 200 μg/mL of proteinase K. Finally, the sample was extracted with phenol: chloroform. The DNA was ethanol precipitated, resuspended in TE pH 8 (10 mmol/L Tris-HCl pH 7.5, 1 mmol/L EDTA) and digested with 100 ng/μL of RNase A for 30 min at 37°C. Purified DNA was subjected to Southern blot analysis. DNA samples were loaded onto 1.3% agarose gels, blotted onto nylon membranes, and probed with a Dig-labeled full-length HBV genome in EasyHyb hybridization solution (Roche, Germany).
For quantitative PCR, 100 μL mouse serum was adjusted to 6 mmol/L MgOAc2 and incubated for 2 h at 37°C with 200 μg/mL of DNase I and 100 μg/mL of RNase A. Following proteinase K digestion, the sample was extracted with phenol: chloroform. HBV DNA levels were analyzed with the Light Cycle real-time PCR system (Roche, Germany), and primer sequences as follows: 5’-TCACAATACCGCAGAGTC-3’ (nt231-248, forward), 5’- AGCAACAGGAGGGATACA-3’ (nt569-552, reverse). The pHBV1.0 vector containing the full-length HBV genome was used as a standard curve to calculate HBV copies per milliliter of serum.
RNA isolation and analysis
For Northern blot analysis, total RNA was extracted from transfected cells with TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Twenty microgram RNA was separated by electrophoresis on a 1% agarose gel and transferred onto nylon membrane, hybridized with a DIG-labeled DNA fragment covering the full-length HBV sequence. The probe was generated with a PCR DIG probe synthesis kit (Roche, Germany). For Northern blot analysis, RNA samples were resolved on MOPS-buffered 1.2% agarose gels containing 1.8% formaldehyde followed by capillary transfer onto nylon membranes and hybridization with a 32P-labeled full-length HBV probe.
For quantitative RT-PCR, approximately 20 mg of liver tissue was obtained from mice for total RNA extraction with RNeasy total RNA kit (QIAGEN, Germany), according to the manufacturer’s protocol. cDNA was synthesized from 2 μg of total RNA with oligo(dT)15 primer in a total volume of 20 μL. Quantitative RT-PCR was performed with the Light Cycle real-time PCR system (Roche, Germany). The PCR primers are as follows, GAPDH: 5’-GTTGTCTCCTGCGACTTCA-3’ (forward), 5’-GGTGGTCCAGGGTTTCTTA-3’ (reverse); HBV: 5’-TCACAATACCGCAGAGTC-3’ (nt231-248, forward), 5’- AGCAACAGGAGGGATACA-3’ (nt569-552, reverse). A standard curve was constructed by the simultaneous amplification of serial dilutions of the expression plasmid encoding HBV used as templates. Target cDNAs were normalized to the endogenous RNA levels of the DAPDH. Quantitative amplification was carried out using the SYBR Green kit (Invitrogen, USA). Gene expression was determined using the relative quantification: ΔΔCT = (CT Target - CT GAPDH) Test - (CT Target - CT GAPDH) Control. CT is the fractional cycle number that reaches a fixed threshold, CTest is the test of HBV, and CControl is the reference control (RNA from control group). ΔCT is the difference between gene expression in treated cells and reference control cells. The fold increase was calculated using 2-ΔΔCT[14].
RESULTS
Inhibitory effect of APOBEC3G on HBV DNA replication in cell culture
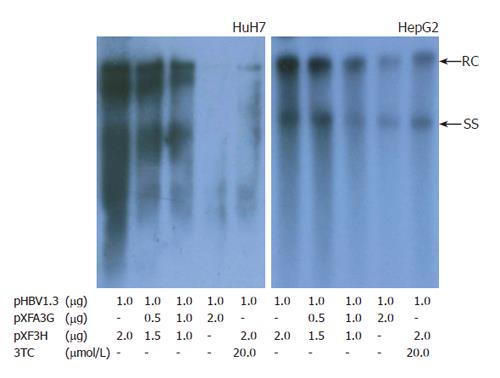
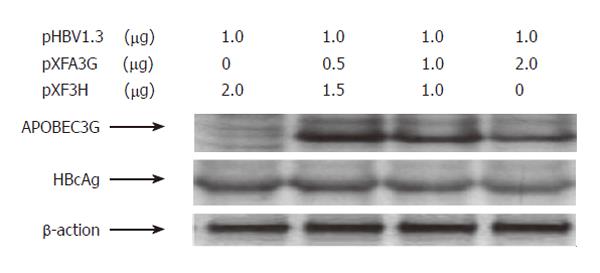
The mammalian hepatoma cells HuH-7 and HepG2 were co-transfected with replication-competent over-length 1.3 fold HBV and various amounts of a CMV-driven expression vector encoding APOBEC3G. In order to determine the efficiency of transfection, Western blot with polyclonal anti-HA and anti-HBV core antibody was used to analyze the expression of APOBEC3G and HBV core protein in co-transfected Huh7 cells. Three days after transfection, core-associated viral DNA was prepared from nuclease-treated cytoplasmic lysates and analyzed with Southern blotting. As shown in Figures 1 and 2, APOBEC3G and HBV core protein were expressed in co-transfected Huh7 cells. However, APOBEC3G reduced the level of replicative HBV intermediates in a dose-dependent manner. This inhibitory effect was also observed in HepG2 cell lines.
Figure 1 Effect of APOBEC3G on HBV replication in cotransfected hepatoma cells.
Cells were treated with lipofectamine 2000 reagents, and viral replicative DNA intermediates were analyzed with southern blotting 3 d after transfection. Numbers at the end of the lines indicate the amount of transfected plasmid DNA in micrograms. HBV, hepatitis B virus; RC, relaxed circular DNA; SS, single -stranded DNA.
Figure 2 Western blot analysis of cytoplasmic extracts from Huh7 cells cotransfected with indicated plasmids.
Inhibition of HBV gene expression by APOBEC3G in cell culture
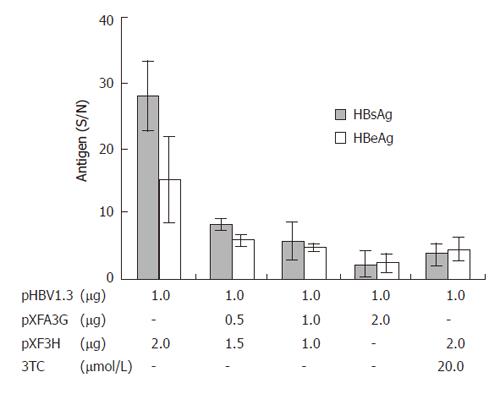
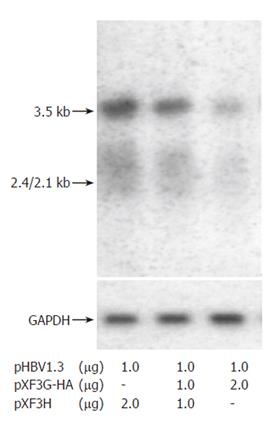
To evaluate the effects of APOBEC3G on HBV gene expression, HuH7 cells were co-transfected with replication competent over-length HBV(pHBV1.3) and various amounts of CMV-driven expression vector encoding APOBEC3G 3 d after transfection. HBsAg and HBeAg levels were determined by ELISA. As shown in Figure 3, there was also a dose dependent decrease in the levels of extracellular production of HBsAg and HBeAg. However, western blot with polyclonal anti-HBV core antibody indicates that the levels of core protein were unaffected by APOBEC3G (Figure 2). In order to examine the effect of APOBEC3G on HBV RNA levels in co-transfected Huh7 cells, total cellular RNA derived from co-transfected Huh7 cells was hybridized with Dig-labeled probe. Northern blot indicated that the RNA levels of 3.5 kb, 2.4/2.1 kb in co-transfected Huh7 cells were significantly suppressed by APOBEC3G (Figure 4).
Figure 3 Effect of APOBEC3G on HBsAg and HBeAg secretion in the media of cotransfected Huh7 cells 3 d after transfection.
HBsAg and HBeAg levels (S/N) were significantly reduced (error bar indicate standard error). Numbers at the end of the lines indicate the amount of transfected plasmid DNA in micrograms.
Figure 4 Effect of APOBEC3G on HBV RNA expression.
Northern blot analysis of HBV transcripts from Huh7 cells co-transfected with indicated plasmids. The same blot was hybridized with a glyceraldehyde phosphate dehydrogenase (GAPDH) DNA probe as an internal control.
Inhibition of HBV replication and gene expression by APOBEC3G in mice
To examine whether APOBEC3G could also display HBV inhibition in vivo, replication-competent HBV plasmid (pHBV1.3) was co-transfected with pXFA3G or pXF3H(control plasmid) to mouse liver by hydrodynamic injection. As shown previously, hydrodynamic injection of pHBV1.3 into mouse resulted in HBV replication in the liver and secretion of viral antigens to the serum. Consistent with in vitro experiments, serum HBsAg and viral DNA in mice which received pHBV1.3 was suppressed dramatically at different time points by APOBEC3G compared with mice without APOBEC3G treatment (Figures 5 and 6). Real time RT- PCR quantification of core-associated HBV RNA in the liver of mice 3 d after injection with APOBEC3G expression plasmid was decreased about 50 times than that of control group (Figure 7).
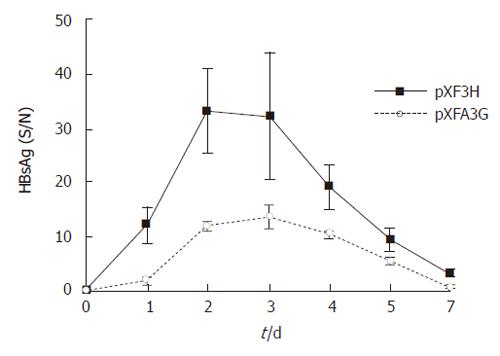
Figure 5 Serum HBsAg levels in APOBEC3G treated mice.
HBsAg levels(S/N) in the sera of BALB/c mice were significantly reduced after treatment with the APOBEC3G expression plasmids (error bar indicate standard error) in the indicated time points, n = 6 per treatment group.
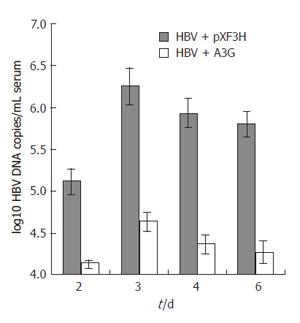
Figure 6 Real time PCR quantification of HBV DNA in the sera of mice treated with APOBEC3G expression plasmid.
BALB/c mice were coinjected with pHBV1.3 vector (10 μg ) and APOBEC3G expression plasmid (10 μg) 3 d after injection. HBV DNA levels (mean ± SD) of sera were determined by quantitative PCR, n = 6 per treatment group.
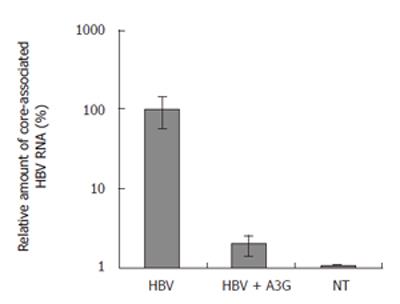
Figure 7 Real time PCR quantification of core-associated HBV RNA in the liver of mice treated with indicated plasmids.
BALB/c mice were coinjected with pHBV1.3 vector (10 μg) and APOBEC3G expression plasmid (10 μg) 3 d after injection. The levels of liver HBV RNA were determined by quantitative RT-PCR, normalized to GAPDH mRNA and are reported as a ratio of HBV mRNA/GAPDH mRNA (mean ± SD). n = 6 per treatment group.
DISCUSSION
This study explored the effects of human APOBEC3G suppressing HBV DNA production in mammalian hepatoma cells as well as in HBV vector-based mouse model. Our data confirm and extend recent results by Turelli et al[10] who reported A3G-mediated inhibition of HBV DNA production in human HuH-7 hepatoma cells. Previous studies suggested that G-to-A hypermutation can influence HBV pathogenesis. Specific G-to-A changes yield HBeAg-negative HBV variants, often isolated from patients with acute fulminant hepatitis, as well as HBV vaccine escape mutants[17,18]. However, there is no evidence so far that APOBEC-induced lethal hypermutation represents an important innate defense mechanism down regulating hepadnavirus production. APOBEC3G-mediated editing did occur but was only detected in a minority of clones produced in transfected HepG2 hepatoma cells[10]. Recently, Rosler et al[19] demonstrated that APOBEC3G rendered HBV core protein associated full-length pregenomic RNA nuclease-sensitive. APOBEC3G-mediated editing of nucleic acids does not seem to represent an effective innate defense mechanism for hepadnaviruses.
During the course of an acute infection, HBV DNA clearance apparently occurs through noncytopathic mechanisms in which IFN-γ and TNF-α play an important role[20,21]. APOBEC3G might participate in this type of antiviral response. Although APOBEC3G is not normally expressed in the liver[10,22], recent study shows that APOBEC3G is induced by interferon-α stimulation in human hepatocytes and human primary monocyte-derived macrophages[16,23]. It is possibly induced by interferon in the course of HBV infection. Several studies have shown that IFNs inhibit HBV replication in vitro in human hepatoma cells and HBV transgenic mice in vivo[24-26]. Currently, IFN-α is approved for treatments of chronic hepatitis B. We speculate that APOBEC3G might be responsible for the anti-HBV action of IFNs in hepatic inflammation. Further analyses will be necessary to determine whether APOBEC3G play roles in the host innate defense against hepatitis viruses in vivo.
The hydrodynamic delivery of nucleic acids in the mouse was described by Liu et al[11], who showed that the vast majority of the injected nucleic acid was delivered to the liver by this technique. Yang et al[27] first demonstrated that hydrodynamic injection of a replication-competent HBV vector resulted in high levels of HBV replication in the livers of injected mice. In the vector-based model, HBV replicates in the liver of immunocompetent mice for 7 to 10 d, resulting in detectable levels of HBV RNA and antigens in the liver and HBV DNA and antigens in the serum. Several reports have documented the use of HBV vector model to examine the in vivo activity of co-HDI administered HBV-targeted unmodified siRNAs[28,29] or vector-expressed short hairpin RNAs in silencing HBV gene expression[30]. Most notably, we have demonstrated in vivo activity of APOBEC3G expression vector via standard intravenous administration. This is the first demonstration of APOBEC3G in vivo activity in a hepatitis animal model with a clinically viable route of administration. Although we have seen a more than 1.5log10 reduction of serum HBV levels with APOBEC3G in an HBV mouse model, however, the potential contribution of toxicity from the APOBEC3G has not been ruled out and needs to be investigated further.
Our work demonstrates APOBEC3G effectively inhibits HBV replication in culture cells and mammalian liver, suggesting that such an approach could be useful in the treatment of HBV infection. However, whether suppression of viral replication by APOBEC3G plays a role during natural HBV infection seems speculative at present, because expression of APOBEC3G in human liver tissue has not yet been shown. Nevertheless, a better understanding of the mechanisms of APOBEC3G action may help identify new therapeutic strategies against chronic hepatitis B.