INTRODUCTION
The human leucocyte antigen (HLA) complex is a highly polymorphic, gene dense region on chromosome 6p21.3 (Figure 1). The complex spans -4 Mb and encompasses at least 130 expressed genes[1,2]. These include the highly polymorphic classical class I and II HLA genes essential for normal lymphocyte function, and a number of other genes with immunoregulatory function. Since the first report of an HLA association with IBD in 1972[3], more than 100 studies have been published investigating the role of HLA genes in determining susceptibility and phenotype of inflammatory bowel disease (IBD). Early data derived from both association and linkage analyses were inconsistent, reflecting the difficulties of inadequate sample size, low resolution typing techniques, poor statistical methodology and a failure to consider disease heterogeneity. However, since the mid-1990's, advances in genotyping and computational technology, combined with improved study design, have been successfully applied to much larger, more accurately characterised cohorts of patients, and this has revealed important clues to molecular pathogenesis of IBD.
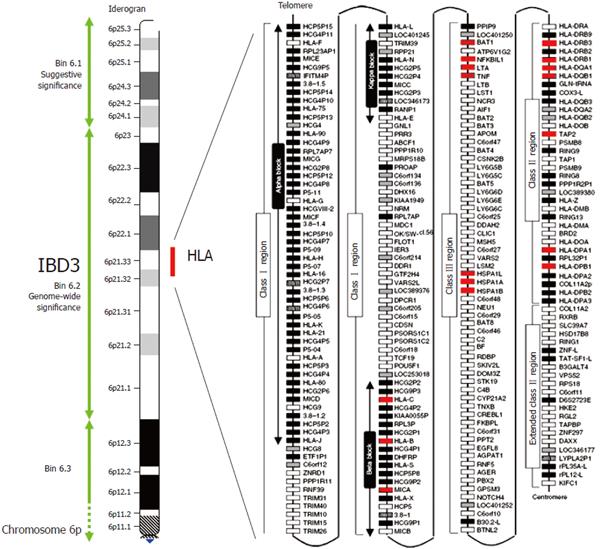
Figure 1 Crohn's disease map of chromosome 6p and the HLA region.
The IBD susceptibility locus IBD3 is here defined by a 34cM chromosomal bin (6.2), which reached genome wide significance for IBD in the genome scan meta-analysis of van Heel et al[10]. The IBD3 region encompasses the ~ 4MB HLA region shown in detail on the right of the figure. White, grey, striped and black boxes show expressed genes, gene candidates, non-coding genes and pseudogenes, respectively. Genes with polymorphisms associate with IBD are shown in red. Modified with permission from Shiina et al[61].
Genome-wide scans have shown consistent evidence of linkage to IBD3 (6p21.1-23), an area which encompasses the HLA complex. This has been demonstrated in several independent studies of both Crohn’s disease[4-8] and ulcerative colitis[5-8], and was recently been confirmed by the IBD Consortium in a large replication study of 733 nuclear families[9]. The importance of this area was further highlighted by a meta-analysis of 10 published genome-wide scans[10]. Notably in this study, IBD3 was the only locus that met genome-wide significance, and provided stronger evidence of linkage than 16p13.1-16q12.2 (IBD1), the locus that contains the susceptibility gene CARD15. Although it is difficult to estimate the importance of this region in determining overall genetic susceptibility, calculations derived from studies of HLA allele sharing within families suggest that this region contributes between 10%-33% of the total genetic risk of Crohn's disease[11] and 64%-100% of the total genetic risk of ulcerative colitis[12].
Interest in the HLA complex in IBD has traditionally focused on association with the classical class II HLA alleles, but recent insights into the biological function of other genes encoded within this region have led investigators to a more diverse exploration of this region.
CLASSICAL HLA CLASS II GENES
The classical class II HLA genes encode cell-surface glycoproteins which are expressed on antigen presenting cells, including dendritic cells and macrophages. Their major role is to present peptides to T cell receptors, as a prelude to T cell activation. There are three types of HLA Class II molecules expressed by a single cell, namely HLA-DR, HLA-DQ and HLA-DP. These are each made up of αβ heterodimers: the β chains are encoded by the DRB1, DQB1 and DPB1 genes respectively, all of which are highly polymorphic. The α chains for DPA and DQA are also highly polymorphic, while the DRA gene is invariant. In addition, gene duplication events have occurred on certain haplotypes producing additional DRB3, 4 and 5 genes. When present, these are expressed at low levels in conjunction with the DRA chain.
The mechanism by which classical HLA class II genes exert their influence in IBD is unclear, although a number of hypotheses have been postulated. Polymorphism in these molecules is concentrated around specific pockets of the binding groove that interact with critical side-chains or 'anchor' residues of a peptide. Thus different HLA molecules may bind preferentially to different peptides, or bind the same peptide with varying affinity. In IBD, cross reactivity (known as "molecular mimicry") may exist between the peptides derived from bacterial luminal flora and from self antigens present in the gut. This may lead to the generation of auto reactive T cells which contribute to disease pathogenesis through either stimulation or inhibition of the immune system. This mechanism is supported by identification of murine MHC-restricted CD4+ T cells reactive to enteric bacterial antigens that are able to induce colitis by adoptive transfer[13].
HLA-DRB1 is the most extensively studied gene in IBD. Convincing evidence of association has been described for a number of alleles, some of which confer risk, whilst others are protective. Many of these were highlighted in a 1999 meta-analysis of 29 association studies published between 1966 and 1998[14]. Since publication of this report, more detailed genotype-phenotype analyses of this locus have been conducted in larger, accurately characterised, patient cohorts leading to the realisation that genetic variation within this region may explain some of the disease heterogeneity of IBD. The most consistently replicated associations are described below.
HLA CLASS II ASSOCIATIONS WITH CROHN’S DISEASE
HLA-DRB1*07 and CARD15 negative ileal Crohn's disease
The most consistently replicated association of Crohn's disease with a common HLA allele is with HLA-DRB1*07. The population frequency of this allele varies between 5%-29% in Europeans and North Americans but is less than 1% in Japanese. The 1998 meta-analysis demonstrated a weak association of HLA-DRB1*07 in unselected patients with Crohn's disease (OR 1.42, CI 1.16-1.74)[14], but three subsequent studies of well-characterised patients from the UK (n = 244)[15], Canada (n = 432)[16] and Spain (n = 210)[17] showed that this association is specifically with patients with ileal involvement (with or without colonic disease). When comparison was made in these 3 studies between patients with, and patients without ileal disease, the Odd's Ratios for ileal disease were 1.5[15], 1.9[16] and 2.6[17] respectively, which are equivalent to those observed for possession of a single CARD15 variant. Importantly the UK and Canadian studies provided evidence for genetic heterogeneity in ileal Crohn's disease, by demonstrating that the association with HLA-DRB1*07 is only present in patients who do not possess one of the three common Crohn's disease associated CARD15 variants[15,16].
Studies in other ethnic groups have not been as detailed. Few Jewish patients were included in the UK or Spanish studies, but data from the Canadian study suggested that this phenotypic association may also be present in this ethnic group, although the relatively small number included prevented this reaching statistical significance[16]. DRB1*07 appears not to be associated with Crohn's disease in the Japanese, although studies have been insufficiently powered to detect a modest effect of such a rare allele.
HLA-DRB1*0103 and Colonic Crohn's disease
HLA-DRB1*0103 is a rare allele with a frequency of less than 2% in European and white North American populations. It is present at a similar frequency in Jews but is absent in the Japanese. A strong association of HLA-DRB1*0103 with ulcerative colitis was identified in 1996[12], but not demonstrated in Crohn's disease until 2000[18], almost certainly a consequence of the disease heterogeneity of Crohn's disease and the low prevalence of this allele. This association has been widely replicated since, although studies in Jewish patients have been inconsistent[16,18]. Association with this allele is observed both on the DRB1*0103-DQB1*0301 and DRB1*0103-DQB1*0501 haplotypes, strongly implicating DRB1*0103 in disease pathogenesis. It is now clear that DRB1*0103 is strongly associated with colonic location, particularly isolated colonic disease[15-17,19]. The Odd's ratios for isolated colonic disease range from 5.1 to 18.5 in non-Jewish caucasoids[15-17]. Data from a recent study suggests that amongst British patients with isolated colonic Crohn's disease, DRB1*0103 is associated with the development of severe disease as defined by the requirement for infliximab or colectomy. Furthermore data from this study suggests this allele may predict time to surgery (personal communication-Laura Hancock). Despite the strength of these associations it should be noted that this allele was present in no more than 32% of patients with isolated colonic disease, indicating that whilst this marker has a high positive likelihood ratio, the negative likelihood ratio limits its clinical application. Association with perianal location and fistulizing behaviour, reported in two studies, probably reflects their clinical association with colonic disease, rather than an independent genetic association[15,17].
HLA-DRB3*0301 - HLA-DRB1*1302
The HLA-DRB3 gene is expressed in less than 50% of Europeans and white North Americans, and has not been extensively studied in IBD. However, a meta-analysis of three studies, each comprising less than 70 patients, supports a positive association of Crohn's disease with DRB3*0301 (OR 2.18, CI 1.25-3.80)[14]. This association has been replicated in a larger cohort of British patients (RR = 2.4)[15], although once again inadequate sample size has prevented investigators determining whether it is due to linkage disequilibrium with another disease associated DRB1 allele, DRB1*1302[14,15], or the HLA class I allele Cw*0802[15]. Although stratification by disease phenotype is limited, data from this latter study, which awaits replication, suggests that this association is most marked in patients with perianal disease.
HLA-DRB1*04 and CARD15 positive ileal disease
Data from the meta-analysis of seven small studies also demonstrated a non-significant increase in the common allele HLA-DRB1*04 in Crohn's disease (OR 1.62 CI 0.73-3.61)[14]. This association is particularly important in Japanese cohorts, in whom the strongest HLA associations are with DRB1*0405, *0410, and the linked DQB1*0401, 0402 alleles[20-22]. Association with DRB1*04 has not been widely observed in European and North American patients. However in the recent Canadian study, a weak association with DRB1*04 was identified in patients with ileal disease RR = 1.7 (1.1-2.5)[16]. It is interesting that this association was stronger in patients possessing one of the three common Crohn's disease associated CARD15 variants, which may indicate an epistatic interaction between this DRB1 allele and CARD15 in determining susceptibility to ileal Crohn's disease[16]. Genotype-phenotype analysis of this allele in the Japanese is awaited.
HLA-DRB1*1501 – Crohn’s disease protection
Much confusion has surrounded associations of Crohn's disease with the serologically defined HLA allele DR2. DR2 includes the molecularly defined alleles HLA-DRB1*1501 and HLA-DRB1*1502, amongst others. The allele frequency of DRB1*1501 varies between 6%-25% in European and white North American populations and 6%-10% in the Japanese. It is negatively associated with Crohn's disease which explains the earlier negative association reported with the serological antigen DR2 (OR 0.83, CI 0.70-0.09) highlighted in the 1999 meta-analysis[14]. This allele appears to confer protection against all subgroups of Crohn's disease, in all ethnic groups including Japanese.
HLA CLASS II ASSOCIATIONS WITH ULCE-RATIVE COLITIS
HLA-DRB1*0103 and severe, extensive ulcerative colitis
The most consistently replicated association of ulcerative colitis in European and American populations is with the rare allele HLA-DRB1*0103. Data from the Stokkers et al[14] meta-analysis, derived from only 3 studies, demonstrated a moderate association in unselected ulcerative colitis patients (OR 3.42 CI 1.52-7.69). Subsequent larger studies have confirmed this association in Spanish[23], North American[18], British[24]and Mexican cohorts[25]. This association is particularly strong in patients with extensive[24-27] or severe disease, as defined by the need for colectomy for failed medical therapy[24,25]. Amongst patients who require colectomy this allele may also be associated with a shorter mean time to surgery[24]. Once again, the frequency of this allele, even in the ulcerative colitis population, is too low to be clinically useful in predicting disease course.
HLA-DRB1*1502
HLA-DRB1*1502 is associated with ulcerative colitis in European[24], North American[18], Japanese[22,28] and Korean[29] populations. Although the background prevalence of this varies considerably, being highest in the Japanese (20%-25%) and lowest in Northern Europeans (less than 1%), the associated relative risk is similar in all populations (2-4.5). This interesting transracial concordance suggests this allele, or a nearby allele, is a true disease causing variant. This association is specifically with HLA-DRB1*1502, rather than HLA-DRB1*1501, and explains the earlier reported association with the serological antigen DR2 (OR 2.00, CI 1.52-2.63) highlighted in the 1999 meta-analysis[14]. These two alleles differ only at amino acid position 86 within pocket 1 of the peptide binding groove. At this position HLA-DRB1*1501 has a valine and DRB1*1502 (and DRB1*0103) a glycine. Limited data suggests HLA-DRB1*1502 is associated with extensive and intractable ulcerative colitis amongst Japanese[30], but not Korean patients[29].
In Japanese[31] and British UC patients[24], association has also been reported with HLA-B*52, the class I allele found in linkage disequilibrium with HLA-DRB1*1502. However, studies have been insufficiently powered to determine which allele represents the primary association due to the highly conserved nature of this haplotype. Interestingly HLA-B*52, but not HLA-DRB1*1502, has also been shown to be associated with colonic Crohn's disease in these populations[15,32], providing further evidence of a shared genetic basis for ulcerative colitis and colonic Crohn's disease.
HLA-DRB1*04-ulcerative colitis protection
The common HLA-DRB1*04 allele is negatively associated with ulcerative colitis in Northern Europeans and Japanese populations, in contrast to the positive association in Crohn's disease. Data from the Stokkers meta-analysis of 15 studies demonstrated an Odd’s Ratio of 0.54 (CI 0.73-3.61) in ulcerative colitis[14]. In the British population, the protective effect of this allele is confined to the most prevalent subtype DRB1*0401, and only then, when present on the two locus haplotype DRB1*0401-DQB1*0301[24]. This suggests that either an interaction between DRB1*0401 and DQB1*0301 is important for disease protection, or, and perhaps more likely, that the true protective polymorphism is found nearby on the associated extended haplotype.
HLA CLASS II ASSOCIATIONS WITH THE EXTRA-INTESTINAL MANIFESTATIONS
A number of HLA associations have been described with the extra-intestinal manifestations of IBD. The majority of data has been derived from small studies from one single centre and should be interpreted with caution until replicated. Small sample size has also hindered attempts at determining the primary association, but the most convincing associations have been described with DRB1*0103[33] in mixed studies of ulcerative colitis and Crohn's disease. Type 1 peripheral arthritis, a migratory pauciarticular large joint arthritis has specifically been shown to be associated with HLA-DRB1*0103, as well as the class I alleles that may be found in linkage disequilibrium, namely B*27 and B*35[33]. In this study, HLA-DRB1*0103 was found in 35% of patients compared to 3% of controls. In patients with recurrent arthritis this association becomes even stronger, being found in 65% of patients[33]. In contrast, type II peripheral arthritis, a chronic, small joint, symmetrical arthritis is associated with HLA-B*44[33]. Uveitis has also been associated with DRB1*0103 and HLA-B*27, and erythema nodosum with the TNF promoter SNP TNF-1031C[34]. However, it is important to note that an increased prevalence of extra-intestinal manifestations has previously been reported in patients with colonic Crohn's disease[35] raising the possibility that the association between DRB1*0103 and the extra-intestinal manifestations may merely reflect the replicated association with colonic disease. Further work is required to clarify this issue.
OBSERVATIONS ON HLA CLASS II ASSO-CIATIONS WITH IBD
A number of important observations should be noted from the reported associations with HLA Class II alleles described above. Firstly, the specific associations with Crohn's disease and ulcerative colitis are different, with the notable exceptions of the shared association of ulcerative colitis and colonic Crohn's disease with HLA-DRB1*0103 and HLA-B*52. Secondly it is clear that multiple alleles, rather than a single allele, may be associated with any specific phenotype. Whilst these associations may merely reflect linkage disequilibrium with a nearby gene, this observation supports peptide presentation as an explanation for these associations, and raises the possibility of a shared disease-associated epitope, as seen in rheumatoid arthritis[36]. Some associated class II alleles do indeed share significant residue changes in the third hypervariable region, which forms part of the antigen binding domain. However these are not shared by all of the associated alleles, suggesting this hypothesis cannot entirely explain these associations. Thirdly, it is not known whether a gene dose effect operates, such that possession of two HLA risk alleles confers additional risk. To date studies in IBD have not been sufficiently powered to conclusively answer this question. Fourthly, whilst many of the associations appear to be robust, it is clear that the penetrance of the genotype is low, and the presence of a risk allele is neither necessary nor sufficient for disease to occur. Finally the reported associations vary with ethnicity and geographical location, reflecting genetic heterogeneity, prevalence of risk alleles in the background population, and population specific patterns of linkage disequilibrium across the HLA. This illustrates the importance of studying ethnically homogenous cohorts in order to prevent population stratification.
OTHER IBD GENES IN THE HLA COMPLEX
In IBD, as with a number of other HLA Class II-associated diseases, there is increasing evidence for more than one susceptibility locus within the HLA complex[37-41]. The evidence is accumulating faster for Crohn's disease than ulcerative colitis, reflecting the focus of recent research efforts towards Crohn's disease. Such a gene(s) might act independently of the class II locus, or interact with disease associated class II alleles on either the cis or trans chromosome. However, the presence of highly conserved haplotypes across such a gene-dense region has made it extremely difficult to dissect out primary associations. In Crohn's disease, it is most likely that a second disease-susceptibility locus is located between the telomeric end of class I, and the central class III regions. Specific associations have been identified with the class I genes HLA-B and Cw, the non-classical MHC class I-related (MIC) genes, MICA and MICB, and the three heat shock protein genes (HSPA1L, HSPA1A, HSPA1B). However most recent interest has focused on a cluster of immunoregulatory genes centred in the HLA class III region, including TNF, LTA, LTB, NFKBIL and BAT1. To date most studies have investigated only a few SNPs in a single gene, chosen because of previously described associations, or for ease of genotyping, rather than their ability to 'tag' a haplotype to comprehensively survey the linkage disequilibrium across the gene. Results have generally been inconsistent, even within phenotypically defined subgroups, and as with other HLA associated diseases, very few studies have fully controlled for linkage disequilibrium with HLA-DRB1. Associations with TNF and the MIC genes are now discussed.
TNF
The most widely studied gene in the HLA class III region is TNF, which encodes a proinflammatory cytokine that is found in increased concentrations in the mucosa, serum and stool of patients with IBD. The ΔARE[42] and TNF-/- mice[43] models provide experimental evidence of the importance of TNF in IBD, and convincing clinical evidence is seen in both Crohn's disease and ulcerative colitis from the dramatic response following infusion of the anti-TNFα monoclonal antibody, infliximab. A large number of TNF promoter polymorphisms have been described, raising the possibility that altered TNF expression may play a role in determining susceptibility to IBD. However the functional significance of individual polymorphisms remains uncertain as TNF regulation is highly cell and context-specific and currently available in vitro systems provide limited insight into how TNF is regulated in vivo.
The most consistent TNF association is with the -857C allele. This is a very common allele: 99% of the healthy caucasoid population possess at least one copy and -85% are homozygote. A modest association with the -857C allele genotype was reported in TDT and case-control analyses from the UK. Relative risks for the homozygote TNF-857CC genotype were 2.4 (CI 1.4-4.0) in ulcerative colitis and 2.4 (CI 1.4-4.2) in Crohn's disease patients lacking the three associated CARD15 variants[44]. This association was replicated in a TDT study from Australia, but unlike the British study, evidence for epistasis was observed between TNF-857C and CARD15 variants[45]. Further evidence for an epistatic interaction was noted in a recent large TDT study from the IBD consortium, in which the TNF-857C allele was shown to be associated with Crohn's disease, but only in CARD15 transmitting families[9]. No association was found in ulcerative colitis, possibly a consequence of the smaller number of ulcerative colitis families included. At present the TNF-857CC genotype has not been found to be associated with a specific Crohn's disease or ulcerative colitis phenotype. An important confounder, which has not been directly investigated by either of these studies, is that the rare, protective -857T allele is the linkage disequilibrium with DRB1*0301[46], an allele shown by several studies to confer protection to Crohn's disease. Thus it is possible that the TNF-857C association simply reflects linkage disequilibrium within this highly complex region.
Modest associations have also been reported with many other polymorphic sites in the TNF promoter. In Crohn's disease these include TNF-1031C[47], TNF-863A[48] and TNF-308A[15,49,50]. The association with TNF-308A appears to be specifically associated with colonic Crohn's disease (with or without colonic disease)[15,49,50], however once again this finding is confounded by the tight linkage disequilibrium across the A1-B8-DR3 ancestral haplotype, the only common haplotype to contain the TNF-308A allele. In small studies TNF-308A has also been shown to be associated with ulcerative colitis[51,52] providing further evidence of shared genetic susceptibility for ulcerative colitis and colonic Crohn's disease. TNF promoter polymorphisms may also predict ulcerative colitis phenotype: In a British study, which requires replication, homozygosity for a common promoter haplotype, defined by the common alleles at each of 6 positions, were more likely to have distal disease that remained distal throughout the period of follow-up[24].
MICA and MICB
The non-classical MHC class I-related (MIC) genes, MICA and MICB, are expressed on the basolateral cell surface of the gastrointestinal epithelium as well as fibroblasts, endothelial cells and dendritic cells. Their expression is markedly increased in response to cellular stress including both viral and bacterial infection[53,54]. MICA and MICB bind to an activating receptor NKG2D, which is expressed on NK cells, CD8 αβ T cells, γδT cells and activated macrophages[55,56]. Interaction between these receptors may directly stimulate cell cytotoxicity as well as providing costimulation for NK and T cell activation.
Exons 2, 3 and 4 of MICA and MICB are highly polymorphic, and 54 MICA alleles have been officially designated to date. The functional significance of these polymorphisms and the nature of selective forces maintaining them are not known, but the presence of a high proportion of non-synonymous substitutions suggests that these polymorphisms are not the result of random mutation alone. Importantly, several MICA alleles have been shown to alter the binding affinity with NKG2D by as much as 30 fold, suggesting they might exert a functional effect on immune activation[57]. Studies involving unselected IBD cohorts from Europe have failed to report consistent associations with MICA or MICB polymorphisms[58-60]. However, in the Japanese, a consistent association has been reported with MICA-A6 and UC, although this does not appear to be independent of the association with the adjacent HLA-B52 allele[31]. In a phenotypically characterised cohort from the UK, MICA*010 and B*1501, which are in linkage disequilibrium, were shown to be associated with perianal Crohn's disease (RR = 2.1)[15], a finding which requires confirmation.
DOES HLA DETERMINATION HAVE A ROLE IN CLINICAL PRACTICE
Clinical observation, supported by molecular and serological data, strongly suggests that IBD is a heterogeneous family of inflammatory disorders. Ultimately, it is hoped that more accurate clinical, serological and molecular definition of these disorders will lead to better understanding of the different biological mechanisms and complex environmental interactions specific to disease subgroups. This would assist clinicians in the prediction of disease course, prognosis, complications and response to therapy. The data reviewed in this report demonstrate that genes in the HLA are important in determining susceptibility and phenotype of Crohn's disease and ulcerative colitis. Although the specific disease causing gene(s) remain unidentified, the strength and consistency of the emerging HLA associations have confirmed the importance of classifying patients both by accurately defined clinical characteristics, and by possession of known genetic risk factors, such as CARD15 disease associated variants.
In both Crohn's disease and ulcerative colitis, the most consistently replicated associations are with the classical class II gene, HLA-DR. In Crohn's disease the strongest associations are observed with subgroups defined by location of disease. No consistent associations have been reported with disease behaviour, a less stable phenotypic characteristic, or with age at diagnosis. At present, there is little information about HLA associations with complications or response to treatment in Crohn's disease. In ulcerative colitis, the strongest HLA associations are seen with overall susceptibility, which is consistently associated with two alleles, DRB1*0103 and DRB1*1502. These alleles vary widely in prevalence across ethnic groups. Fewer subgroup associations have been identified than with Crohn's disease, but the association with DRB1*0103 is particularly strong in patients with extensive or severe disease. However, the low sensitivity and specificity of these associations currently preclude their use as a tool in diagnosis of either Crohn's disease or ulcerative colitis. For the same reason, it is unlikely that the use of HLA markers alone will be useful in disease screening of asymptomatic, genetically high risk individuals.
In addition to the interest in using biomarkers to predict IBD susceptibility, there is also considerable interest in using molecular and serological markers to assist in the discrimination of Crohn's disease from ulcerative colitis. Currently, markers in the HLA cannot be reliably used for this purpose. Indeed the shared association of DRB1*0103 and B*52 with ulcerative colitis and colonic Crohn's disease suggests the presence of at least one shared HLA susceptibility factor, providing a tantalising clue as to the potential molecular basis for the definition of colonic inflammation. Such shared HLA factors may explain in part why both forms of IBD can coexist in a family at a frequency greater than expected by chance. The apparent differential effect of HLA-DRB1*0401, which is a susceptibility allele in Crohn's disease and a protective allele in ulcerative colitis is interesting, but weak, population specific, and neither sufficiently sensitive or specific to be clinically useful.
In the near future, knowledge of an individual's HLA genotype is likely to be most useful in the prediction of disease course in patients with an established diagnosis of Crohn's disease or ulcerative colitis. However, at present, the sensitivity and specificity of even the most robust associations limit their application in isolation in this clinical context. Integration of an individual's HLA genotype into a panel of other genetic and serological markers may improve the sensitivity and specificity, although ultimately this is likely to require the identification of the primary IBD genes within this complex region.
In common with virtually all other HLA associated complex diseases, the HLA susceptibility gene(s) for IBD remain elusive. This is a consequence of the obstacles that challenge the mapping of disease genes within this region, including the high gene density and degree of polymorphism; the complex haplotype structure and patterns of linkage disequilibrium; the relatively small relative risks conferred by disease associated alleles; and the likely clustering of more than one IBD susceptibility gene, either within the HLA, or within the surrounding IBD3 locus.
S- Editor Pan BR E- Editor Liu Y