Published online Mar 14, 2006. doi: 10.3748/wjg.v12.i10.1536
Revised: September 1, 2005
Accepted: October 9, 2005
Published online: March 14, 2006
AIM: To detect the new serum biomarkers for colorectal cancer (CRC) by serum protein profiling with surface-enhanced laser desorption ionisation - time of flight mass spectrometry (SELDI-TOF MS).
METHODS: Two independent serum sample sets were analysed separately with the ProteinChip technology (set A: 40 CRC + 49 healthy controls; set B: 37 CRC + 31 healthy controls), using chips with a weak cation exchange moiety and buffer pH 5. Discriminative power of differentially expressed proteins was assessed with a classification tree algorithm. Sensitivities and specificities of the generated classification trees were obtained by blindly applying data from set A to the generated trees from set B and vice versa. CRC serum protein profiles were also compared with those from breast, ovarian, prostate, and non-small cell lung cancer.
RESULTS: Mass-to-charge ratios (m/z) 3.1×103, 3.3×103, 4.5×103, 6.6×103 and 28×103 were used as classifiers in the best-performing classification trees. Tree sensitivities and specificities were between 65% and 90%. Most of these discriminative m/z values were also different in the other tumour types investigated. M/z 3.3×103, main classifier in most trees, was a doubly charged form of the 6.6×103-Da protein. The latter was identified as apolipoprotein C-I. M/z 3.1×103 was identified as an N-terminal fragment of albumin, and m/z 28×103 as apolipoprotein A-I.
CONCLUSION: SELDI-TOF MS followed by classification tree pattern analysis is a suitable technique for finding new serum markers for CRC. Biomarkers can be identified and reproducibly detected in independent sample sets with high sensitivities and specificities. Although not specific for CRC, these biomarkers have a potential role in disease and treatment monitoring.
- Citation: Engwegen JY, Helgason HH, Cats A, Harris N, Bonfrer JM, Schellens JH, Beijnen JH. Identification of serum proteins discriminating colorectal cancer patients and healthy controls using surface-enhanced laser desorption ionisation-time of flight mass spectrometry. World J Gastroenterol 2006; 12(10): 1536-1544
- URL: https://www.wjgnet.com/1007-9327/full/v12/i10/1536.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i10.1536
Colorectal cancer is the third most common cause of cancer-related death in both men and women, accounting for about 10% of all cancer deaths annually. When diagnosed and treated early, the overall 5-year survival rate is around 90%. However, most patients present with locally advanced or metastasised disease at the time of diagnosis, or develop metastasis during follow-up. Suitable tumour markers will facilitate colorectal cancer detection, determination of prognosis, and disease and therapy evaluation.
However, currently used non-invasive methods, such as measurement of serum carcinoembryonic antigen (CEA) levels, faecal occult blood testing and faecal DNA analysis, have low sensitivities and/or specificities for colorectal cancer[1-4]. Although CEA is currently the best available marker for follow-up of resected colorectal cancer and monitoring of chemotherapy, its use to determine eligibility for adjuvant therapy or its routine use as a single parameter for treatment monitoring has significant clinical limitations[5,6].
Detection of so-called biomarker proteins in serum may lead to new and better tumour markers for colorectal cancer. The proteome, contrary to the genome, is not a static parameter: it reflects not only the presence of active or inactive (mutated) genes, but also their extent of expression at a specific time point. In addition, the proteome reflects all proteins and peptides that may rise from only one gene, i.e. different cleavage products and proteins with different post-translational modifications. Both characteristics allow a more detailed evaluation of a disease status using the human proteome.
Protein profiling in complex biological matrices has become more easily achievable with the Surface-Enhanced Laser Desorption Ionisation (SELDI) ProteinChip technology in combination with a time of flight (TOF) mass spectrometer. This is a relatively new technique, which lacks the disadvantages of 2D-gel-electrophoresis for proteomic research in that it has high sensitivity in the low molecular weight range and high throughput capability, and proteins with extreme characteristics (highly hydrophobic, acidic or basic) can be analysed more easily[7,8]. With this technique, whole serum is applied to protein chips with different chromatographic affinities in a suitable binding buffer. Selectively bound proteins are retained on the surface and non-selectively bound proteins are washed off. In the mass spectrometer, a laser desorbs the bound proteins from the chip surface, which are subsequently detected in the TOF analyser by their respective mass-to-charge ratios (m/z). Since a whole pattern of proteins is analysed, more than one biomarker can be detected. Combination of several of these biomarkers for the evaluation of a patient’s status may result in enhanced sensitivities and specificities.
SELDI-TOF MS has already been applied to several forms of cancer, including breast, ovarian, prostate, and lung cancer[9-12]. In the obtained protein profiles, proteins with high sensitivity and specificity for disease have been detected. For colorectal cancer, a discriminative protein of 12*103 Da has been found in tumour cell lines, the identity of which was prothymosin-alpha[13]. Comparing epithelial colorectal carcinoma cells with normal tissue, 3.48×103-, 3.55×103- and 3.6×103-Da proteins were found to be increased in cancer tissue[14]. In Asian patients with colorectal cancer and healthy controls, discriminating serum protein profiles have been recently reported, m/z 5911, 8922, 8944 and 8817 being the most important biomarkers[15-18]. These results were obtained in a single sample set and the reported sensitivities and specificities resulted from cross-validation within this single set. In addition, the identities of the reported biomarker proteins remain unknown.
The objective of this study was to detect biomarker proteins for colorectal cancer in serum using SELDI-TOF MS, and to validate these with an independent sample set. In addition, we aimed at identifying any found biomarkers so that further insight into the pathological processes involved in colorectal cancer can be obtained.
Two independent serum sample sets were analysed for their protein profiles on different occasions. The first set consisted of samples from 40 patients with colorectal cancer (all Dukes’ D) and 49 healthy controls. The second set consisted of samples from 37 patients with colorectal cancer (1 Dukes’ A, 2 Dukes’ B, 12 Dukes’ C, 17 Dukes’ D, 5 unknown) and 31 healthy controls. For comparison of colorectal cancer protein profiles with those from other tumour types, a third sample set consisting of serum samples from 8 non-small cell lung cancer (NSCLC) patients (stage III and IV), 10 breast cancer patients (stage II and III), 10 prostate cancer patients (stage I-IV), and 10 ovarian cancer patients (stage I-IV) was analysed. All serum samples were obtained from the serum bank at the Netherlands Cancer Institute, where they were stored at -30 °C until analysis. Sample collection was performed after taking individuals’ informed consent under approval of the Institutional Review Board. Samples were drawn before surgery or chemotherapy was started, except for 9 patients with metastatic disease in sample set B who had already had surgery.
Protein profiling was performed using SELDI-TOF MS (Ciphergen Biosystems Inc., Freemont, CA, USA). Several chromatographic chip surfaces and binding conditions were screened for discriminative m/z values between colorectal cancer patients and healthy controls. The most discriminating peaks were seen on CM10 chips, a weak cation exchange chip, which contains anionic carboxylate groups that bind positively charged proteins in serum. Best results were obtained using a sodium phosphate binding buffer (pH 5) and a 500 mL/L solution of sinapinic acid (SPA; Ciphergen Biosystems) in 500 mL/L acetonitrile (ACN) + 5 mL/L trifluoracetic acid (TFA) as energy absorbing molecule.
All serum samples were denatured by adding 180 µL of 9 mol/L urea, 20 g/L CHAPS, 10 g/L DTT (all from Sigma, St. Louis, MO, USA) to 20 µL of serum. CM10 chips were assembled in a 96-well format bioprocessor (Ciphergen Biosystems) which can hold twelve 8-spot protein chips. During all steps of the protocol, the bioprocessor was placed on a platform shaker at 350 r/m. Chips were equilibrated twice with 200 µL of binding buffer consisting of 20 mmol/L sodium phosphate (Sigma) buffer (pH 5) with 1 g/L TritonX-100 (Sigma) for 5 min. Subsequently, 180 µL of binding buffer and 20 µL of denatured sample were applied to the chip. Sample allocation was at random. Incubation was set to 30 min. After binding, the chips were washed twice for 5 min with binding buffer, followed by two 5-min washes with binding buffer without TritonX-100. Lastly, chips were rinsed with deionised water. After air-drying, two times 1 µL of the SPA was applied to the spots.
Protein chips were analysed using the PBS-IIC ProteinChip Reader (Ciphergen Biosystems). Data were collected between 0 and 200 000 Da. Data collection was optimised for detection of discriminating peaks, resulting in an average of 65 laser shots per spectrum at laser intensity 150 and detector sensitivity 8, and laser focusing at 3000 Da. M/z values for the detected proteins were calibrated externally with a standard peptide mixture (Ciphergen Biosystems) containing [Arg8] vasopressin (1 084.3 Da), somatostatin (1 637.9 Da), dynorphine (2 147.5 Da), ACTH (2 933.5 Da), insulin β-chain (bovine) (3 495.9 Da), insulin (human recombinant) (5 807.7 Da), and hirudin (7 033.6 Da).
Data were analysed with ProteinChip Software package, version 3.1 (Ciphergen Biosystems). For each sample set, all acquired spectra were compiled and analysed as a whole. Spectra were baseline subtracted and normalised to the total ion current between 1 500 and 200000 Da. For validation of either sample set, the normalisation factor from the training set was applied to the spectra of the validation set. The Biomarker Wizard (BMW) software application (Ciphergen Biosystems) was used to autodetect m/z peaks with a signal-to-noise ratio of at least 5. Peak clusters were completed with peaks with a signal-to-noise ratio of at least 2 in a 0.5% cluster mass window. For validation purposes, peak clusters of the training set were applied in the validation set. Group differences were calculated with the same application, comparing mean intensities of all detected peaks between groups with non-parametric statistical tests. P values less than 0.01 were considered statistically significant.
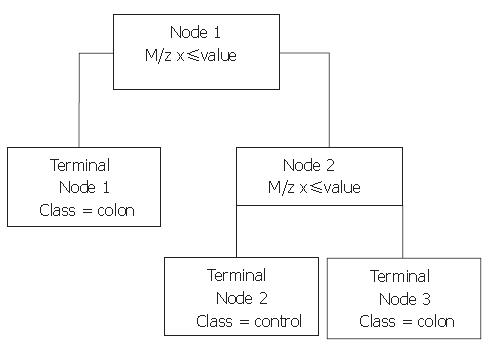
Next, Biomarker Patterns Software (BPS; Ciphergen Biosystems) was used to generate classification trees from the BMW files. A classification tree is built of nodes with an m/z value and a cut-off value for the peak intensity. An example of such a tree is shown in Figure 1. When an analysed spectrum has a peak intensity at the specified m/z below the cut-off value, the sample is placed in the left tree branch. Otherwise, it is placed to the right and its peak intensity at the next m/z value is evaluated. Peaks that result in a maximum separation of the two groups, with a minimum of misclassification are chosen for the nodes. The branch consists of new nodes with an m/z value until a final classification can be made for the spectrum: originating from a colorectal cancer patient or from a healthy control. For every tree, the BPS performs a ten-fold cross-validation in the tree building process, in which ten times another tenth of the data set is used for testing of the tree and these results are combined to yield a cross-validation sensitivity and specificity as a measure for the tree’s discriminative power. However, to obtain a more realistic sensitivity and specificity, classification trees built with one sample set as the training set were validated with the blinded data from the remaining set. In addition, both sample sets were combined to form a training set with two thirds of all samples keeping a random third behind in the tree building process for independent validation afterwards.
Biomarkers detected in the profiling experiments were purified by fractionation of denatured serum on QhyperD beads (Biosepra; Ciphergen Biosystems), a strong anion exchange surface, with decreasing buffer pH. Subsequently, fractions containing the markers were concentrated on microcon YM-50 filters (Millipore, Billerica, MA, USA) and eluted with increasing concentrations of ACN + TFA (1: 0.001, v/v). The purification process was monitored by profiling each fraction on NP20 chips, containing a non-selective, silica chromatographic surface. Relevant eluates were evaporated and redissolved in loading buffer for SDS-PAGE. Gel electrophoresis was performed on Novex NuPage gels (Invitrogen, San Diego, CA, USA). Gels were stained using colloidal Coomassie staining (Simply Blue; Invitrogen) and protein bands of interest were excised and collected for either passive elution, followed by in-solution digestion or in-gel digestion with trypsin (Promega, Madison, WI, USA).
For passive elution, bands were washed twice with 300 mL/L ACN + 100 mmol/L NH4HCO3 (Sigma), followed by dehydration in ACN. Samples were heated at 50 °C and then eluted with 15 µL of formic acid/ACN/isopropanol/deionised water (4.5:3:1:1.5, v/v) under sonification for 30 min. The eluate was left for 3 h at room temperature before profiling on NP20 chips. Then, the eluate was left overnight. In-solution digests were obtained by evaporation of the supernatant in a SpeedVac, resuspending it in 20 mg/L trypsin in 100 mL/L ACN + 25 mmol/L NH4HCO3 and incubation for 4 h at room temperature.
In-gel digestion was performed after washing the excised band with methanol/acetic acid/deionised water (4:1:5, v/v) twice, followed by a wash with 300 mL/L ACN + 100 mmol/L NH4HCO3. Samples were dried on a SpeedVac and immersed in a 20 mg/L-solution of trypsin in 100 mmol/L NH4HCO3. Digestion was allowed for 12 h at room temperature. All tryptic digests were profiled on NP20 chips, using 1 µL 200 g/L CHCA (Ciphergen Biosystems) solution in 500 mL/L ACN + 5 mL/L TFA as matrix.
Peptides in the resulting digest were investigated with the MASCOT and ProFound search engines (http://www.matrixscience.com; http://prowl.rockefeller.edu/profound_bin/WebProFound.exe), using the Swiss-Prot and NCBI databases, respectively. Data were searched against the Homo sapiens subset of the database, defining fixed modification of the cysteine residues with propionamide and variable modification of methionine residues (oxidation). Peptide mass tolerance of the average MH+ masses was 0.5 - 3 Da; the number of tryptic miscleavages allowed was 1 or 2.
Identification of proteins was confirmed either by immunoassay on protein A beads (Biosepra; Ciphergen Biosystems) with an appropriate antibody (Abcam Ltd, Cambridge, UK), or by sequencing of the most important peptides in the tryptic digest with tandem MS on both a Q-TOF™ II, (Micromass Ltd, UK) and a QSTAR™ (AB/Sciex, Foster City, CA, USA), both equipped with a PCI 1000 interface (Ciphergen Biosystems). For the immunoassay, beads were loaded with antibody in phosphate-buffered saline (PBS, Sigma), and washed twice with PBS, followed by a 30-min incubation with whole serum, 5 subsequent washes with PBS and one with deionised water. Finally, bound proteins were eluted using 1 mol/L acetic acid and the eluate was profiled on NP20 chips.
Serum CEA was quantified using an electrochemiluminiscence immunoassay on a Modular analytics E170 analyser (Roche Diagnostics, Mannheim, Germany). A cut-off value of 5 µg/L was employed. Levels of the acute phase reactants C-reactive protein (CRP) and transferrin (TRF) were assessed by a near infrared particle immunoassay and a turbidimetric immunoassay, respectively, using the Beckman Synchron LX20 analyser (Beckman-Coulter Inc., Fullerton, CA, USA). CRP levels below 8 mg/L were considered clinically normal, as are TRF levels between 2.1 and 3.8 g/L. All statistical analyses for these data were performed with SPSS, version 11.0 (SPSS Inc., Chicago, IL, USA).
In the first and second sample set respectively, 15 and 6 proteins of which the expression differed in colorectal cancer patients compared to healthy controls (P < 0.01) were detected with the BMW application. Peaks below 2 000 Da were discarded, as they result mainly from the SPA matrix. In either sample set, m/z values of 3.2*103, 3.3*103, 6.4×103, 6.6×103, 6.8×103, and 28×103 were differentially expressed. Expressions of m/z 2.7×103, 3.1×103, 4.2×103, 4.3×103, 4.5×103, 8.0×103, 8.9×103, 14×103, and 16×103 significantly differed only in sample set A.
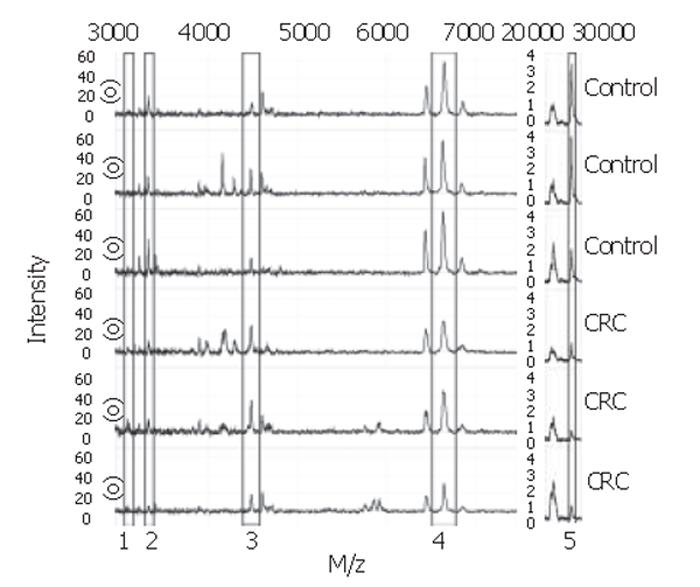
With the BPS several classification trees were built. Tree characteristics of the best-performing trees with accompanying sensitivities and specificities are described in Table 1. Tree I and II were generated from sample set A, tree III and IV from sample set B, and tree V and VI from the combination of sample sets A and B. Tree sensitivities and specificities for trees I to IV were obtained using the second sample set as validation set. For trees V and VI, the sensitivity and specificity were calculated by randomly choosing one third of all data to be excluded from the tree building process for use as validation data afterwards. As shown in Table 1, m/z 3.3×103 was the most frequently observed classifier among these best trees. When removing this classifier from the tree-building model, equally- or better-performing trees were seen with m/z 28×103 as main classifier (trees II and VI). Other biomarkers used in the trees include m/z 3.1×103, 4.5×103, and 6.6×103. Of these, m/z 3.1×103 and 4.5×103 were more abundant in colorectal cancer serum samples compared to healthy controls, whereas the others were less abundant. Parts of representative MS-spectra for patients and controls are shown in Figure 2.
| Tree | Included m/z’s (×103 Da): cut-off intensity values and class assignment | Sensitivity (%) | Specificity (%) | |||
| Node 1 | Node 2 | Node 3 | Node 4 | |||
| I | 3.3 ≤ 15.035 Colorectal cancer | - | - | - | 77.8 | 73.3 |
| II | 28 ≤ 1.558 Colorectal cancer | 4.5 ≤ 29.791 to Node 3 | 3.1 ≤ 9.866 to Node 4 | 6.6 ≤ 33.233 Colorectal cancer | 77.8 | 73.3 |
| III | 3.3 ≤ 12.757 Colorectal cancer | - | - | - | 66.7 | 83.3 |
| IV | 3.3 ≤ 12.757 Colorectal cancer | 28 ≤ 1.285 Colorectal cancer | - | - | 75.0 | 83.3 |
| V | 3.3 ≤ 12.981 Colorectal cancer | 4.5 ≤ 28.599 Healthy control | - | - | 84.2 | 83.3 |
| VI | 28 ≤ 1.529 Colorectal cancer | 4.5 ≤ 28.577 to Node 3 | 6.6 ≤ 44.685 Colorectal cancer | - | 89.5 | 88.9 |
To determine the selectivity of the observed protein profiles for colorectal cancer when compared to other cancer forms, additional samples from patients with other tumours were analyzed. This third sample set was analyzed concomitantly with 17 of the previously analyzed samples from colorectal cancer patients (10 from sample set A, 7 from B) and 20 previously analyzed control samples (10 from either sample set) using the same assay procedures.
When examining peak intensity differences between cancer patients and healthy controls by means of the Biomarker Wizard application, most of the biomarkers for colorectal cancer were found to be discriminative for other cancer forms as well (Table 2). Except for breast cancer samples, m/z 3.3×103, 6.6×103, and 28×103 were markedly less abundant in all types of cancer compared to the control samples (P < 0.01). For these tumour types, mean peak intensities were not significantly different from those for colorectal cancer, independent of patient characteristics (data not shown). There was a tendency for a significant increase of m/z 4.5×103 in ovarian and prostate cancer (P < 0.05). In breast cancer patients, no significance was reached for peak intensity differences of m/z 3.3×103, and 4.5×103 (P = 0.10 and P = 0.54, respectively). However, m/z 3.1×103 was found to be significantly increased only in breast cancer samples (P = 0.0011), but not in other cancer types.
| Group | 3.1×103 Da | 3.3×103 Da | 4.5×103 Da | 6.6×103 Da | 28×103 Da | 5.9×103 Da | ||||||
| Intensity | P (×10-3) | Intensity | P (×10-3) | Intensity | P (×10-3) | Intensity | P (×10-3) | Intensity | P (×10-3) | Intensity | P (×10-3) | |
| HC (n = 20) | 5.51 (2.4) | 14.9 (4.73) | 20.6 (9.16) | 52.1 (7.4) | 2.73 (1.25) | 7.26 (4.23) | ||||||
| CRC (n = 17) | 7.95 (4.64) | 211 | 9.05 (3.81) | 1.11 | 25.5 (5.99) | 12.4 | 37.6 (9.17) | 0.0388 | 1.44 (0.65) | 0.801 | 10.84 (7.66) | 161 |
| BC (n = 10) | 8.87 (2.59) | 1.13 | 11.51 (3.4) | 103 | 19.44 (8.16) | 538 | 42.7 (7.58) | 5.59 | 1.44 (0.59) | 3.69 | 4.51 (2.55) | 43.0 |
| NSCLC (n = 8) | 5.00 (2.93) | 508 | 7.93 (3.38) | 2.27 | 25.1 (7.93) | 93.3 | 30.9 (11.2) | 0.205 | 0.942 (0.45) | 0.450 | 1.91 (2.13) | 0.305 |
| OC (n = 10) | 7.29 (3.77) | 218 | 9.89 (2.52) | 3.69 | 27.7 (8.67) | 27.8 | 40.6 (8.65) | 2.07 | 1.31 (0.45) | 0. 968 | 2.98 (1.47) | 1.32 |
| PC (n = 10) | 7.65 (3.25) | 131 | 9.59 (4.35) | 7.21 | 29.6 (10.1) | 18.4 | 36.5 (11.5) | 0.967 | 1.26 (0.56) | 1.35 | 2.73 (2.01) | 1.86 |
A decision tree combining data from all tumour types was built with the BPS. Although most of the earlier observed biomarkers were discriminative for all other cancer forms as well, 76% of samples from colorectal cancer patients could be correctly distinguished from those of other cancers based on a classifier peak at m/z 5.9×103. In samples from colorectal cancer, peak intensities for this m/z were slightly higher compared to the controls, whereas they were significantly lower than the controls in the other cancers (Table 2). In addition, data from the other tumour types were applied to the trees in Table 1. For all trees, more than 89% of patients with other cancers than colorectal cancer were classified as having colorectal cancer, except for tree VI, in which this was 78.4%.
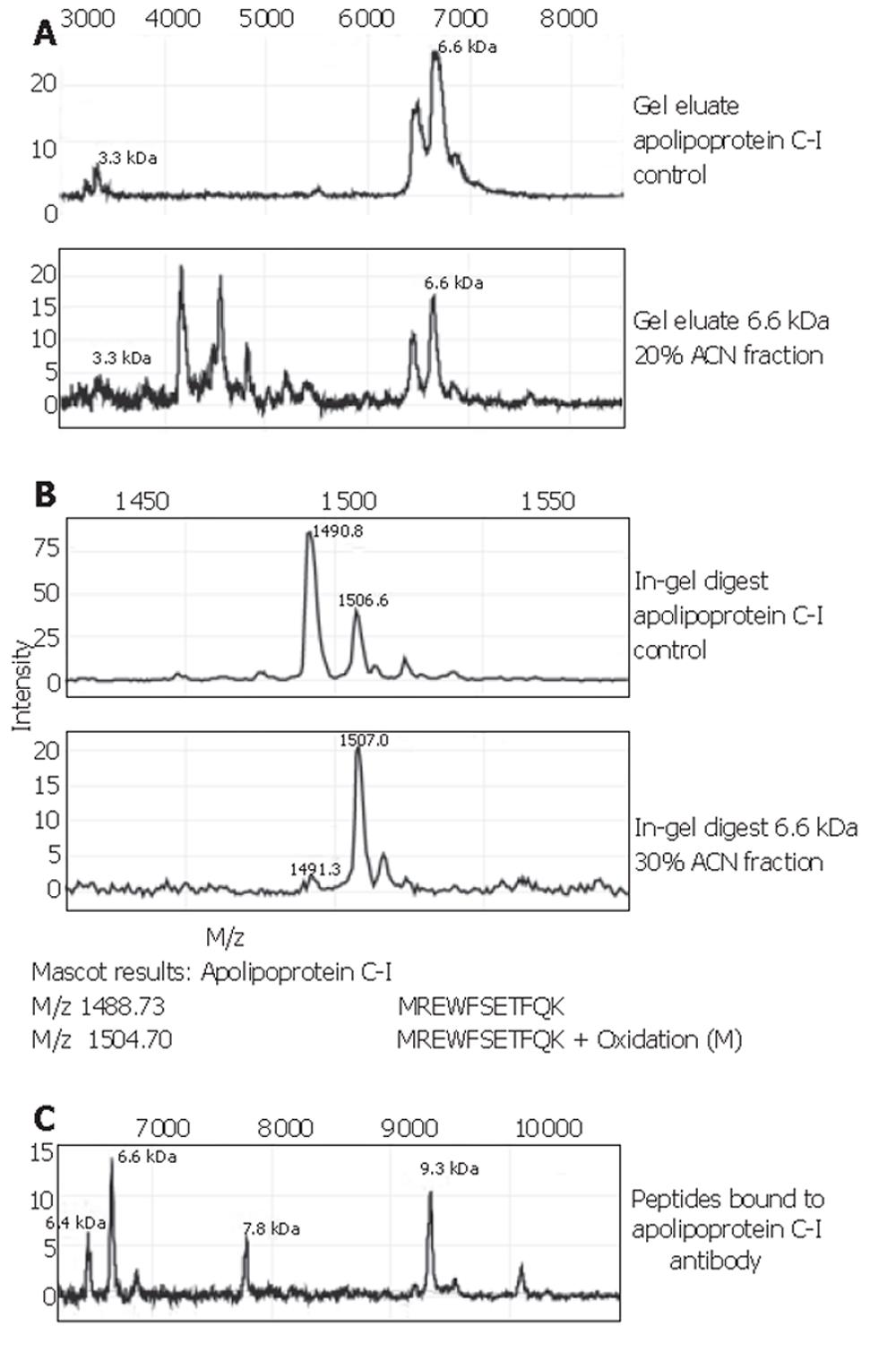
Fractionation of whole serum from colorectal cancer patients and controls resulted in elution of the 6.6×103-Da marker mainly in the flowthrough (pH 9), and the 28*103-Da marker in the pH 4 fraction. Following concentration on YM-50 filters, the 6.6×103-Da marker was seen in the 200 mL/L- and 300 mL/L-ACN eluates mostly, more purified from surrounding masses in the latter. The 28×103-Da marker was present in the filter wash (1 mL/L TFA).
SDS-PAGE of selected eluates was performed on a 120 g/L Bis-Tris gel for the 28×103-Da marker and a 180 g/L Tris-glycine gel for the 6.6×103-Da marker. For this marker, both the 200 mL/L- and 300 mL/L-ACN eluates were placed on gel. A clear band was seen for the 28×103-Da protein, which was divided in half for both passive elution and in-gel digestion. For the 6.6×103-Da marker, a number of faint bands was seen in the 6000-Da region, in both the 200 mL/L- and 300 mL/L-ACN eluates. All were excised. Bands from the 200 mL/L-ACN eluate were subjected to passive elution, and from the 300 mL/L-eluate to in-gel digestion. Profiling of gel eluates on NP20 confirmed the masses to be indeed 6.6×103 and 28×103 Da.
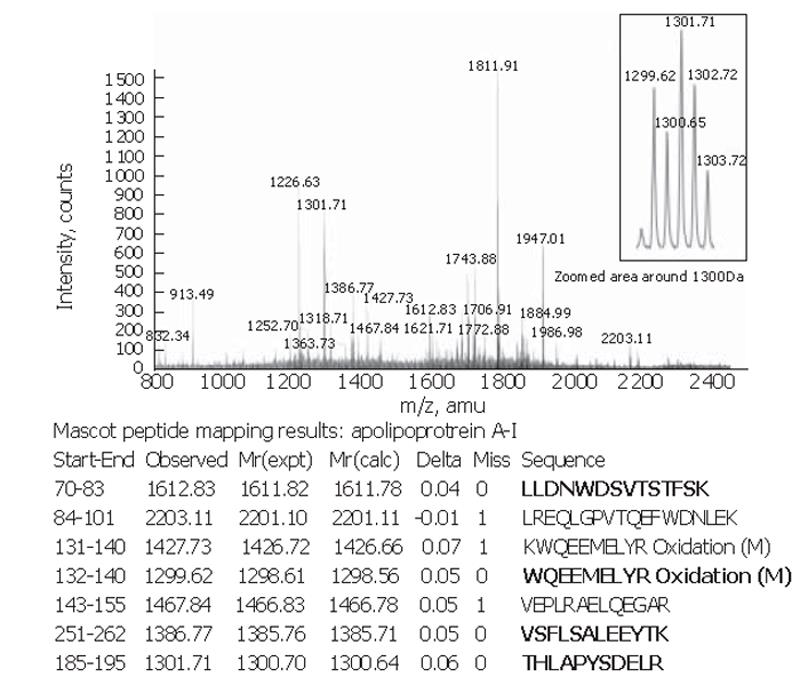
Peptide mapping results revealed the identity of the 28×103-Da marker to be apolipoprotein A-I. The theoretical mass of this protein is 28 078.62 Da in the SwissProt database and its pI = 5.27. The apolipoprotein A-I identity was confirmed by tandem MS of the 1 299.62-, 1301.71-, 1612.83- and 1386.77-Da peptides in the tryptic digest (Figure 3).
The 6.6×103-Da marker was identified as apolipoprotein C-I, with a theoretical mass of 6 630.58 Da and pI=7.93. Spectra of this identification are shown in Figure 4. Confirmation of the apolipoprotein C-I identity was performed on protein A beads using a goat apolipoprotein C-I polyclonal antibody. The eluate’s MS-spectrum clearly showed a large peak at 6.6×103 and another prominent peak at 9.3×103. The mass of the latter peak corresponded to that of apolipoprotein C-I precursor. In addition, the passive elution of the apolipoprotein C-I control (Figure 4A) showed a peak at 3.3×103. The 3.3×103-Da biomarker found in our sample sets A and B, as well as the set combined with other tumour types, consistently showed a high correlation with the 6.6×103-Da one: the ratio of their peak intensities was quite constantly ranging between 3.5 and 4.0 (Table 2). This supports the fact that the observed 3.3×103-Da marker is actually a doubly charged artefact of the 6.6×103-Da protein. The 3.1×103-Da protein was lost during the purification process and was therefore directly sequenced on-chip. It was identified as a 27-amino acid N-terminal fragment of albumin with sequence: DAHKSEVAHRFKDLGEENFKALVLIAF. The identity of the 4.5×103-Da protein is still under investigation.
Evaluation of the extent of a possible acute phase reaction was done by measurement of CRP and TRF levels. Mean TRF levels in the patient and control group were 2.37 g/L (range, 1.20-3.60 g/L) and 2.59 g/L (range, 1.90-4.00 g/L), respectively (P=0.037, non-parametric Mann-Whitney U test). Mean CRP levels in either group were 29.0 mg/L (range, 0 - 213 mg/L) and 3.70 mg/L (range, 0 - 29.2 mg/L) (P < 0.000, non-parametric Mann-Whitney U test). Although there was a significant difference in the levels of these acute phase reactants in the patients and controls, the mere presence of an acute phase response was not a good predictor for colorectal cancer: the sensitivities of CRP and TRF were 51.9% (40/77) and 22.1% (17/77), respectively, using the clinical cut-off values [specificities 88.8% (71/80) and 96.3% (77/80), respectively]. In addition, CRP and TRF levels were included in the BMW data files for the tree-building process with the BPS, in order to evaluate their capability to distinguish colorectal cancer patients and healthy controls. Neither CRP, nor TRF concentrations were as good a classifier as the m/z values in the generated trees.
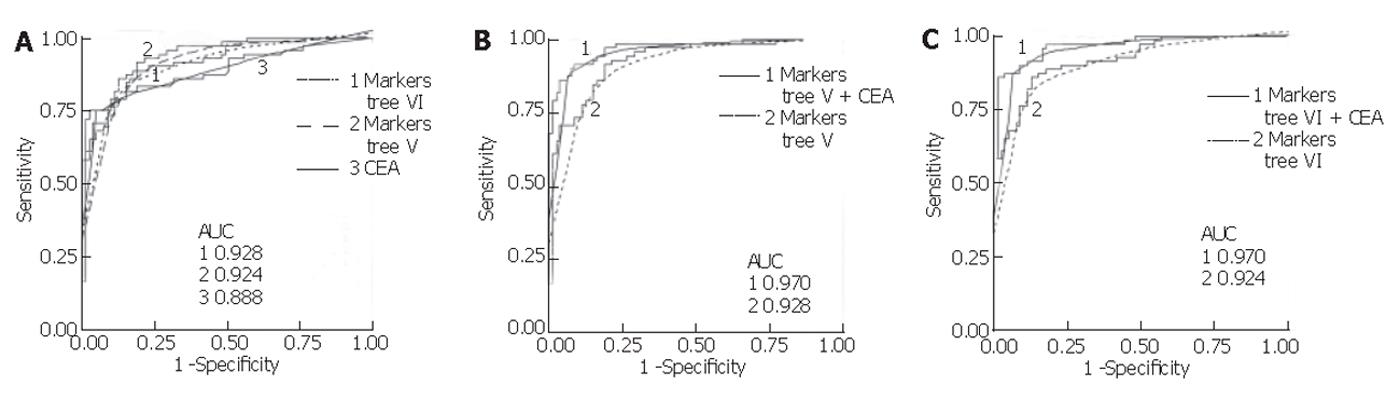
Mean serum CEA in the colon group was significantly higher (mean 326.2 µg/L, range <1-9 452) compared to the control group (mean 2.23 µg/L, range <1-18.98) (P < 0.001, non-parametric Mann-Whitney U test). Using a cut-off value of 5 µg/L, its sensitivity and specificity were found to be 75.3% and 95.0%, respectively, for all samples combined. This sensitivity was lower than that for the proteins in the classification trees generated with all samples (V and VI), but the specificity of CEA in this population was higher. Assessing CEA sensitivity in the total sample set according to colorectal cancer stage, using the 5 µg/L cut-off, resulted in correct classification of 0 of 1 Dukes’ A, 0 of 2 Dukes’ B, 3 of 12 (25.0%) Dukes’ C, and 51 of 57 (89.5%) Dukes’ D. In comparison, using the total sample set and the trees generated with the sets (V and VI), 1 of 1 Dukes’ A, 1 of 2 Dukes’ B, 11 of 12 (91.7%) Dukes’ C, and 47 of 57 (82.5%) Dukes’ D were correctly classified. In addition, logistic regression was performed for CEA and the markers from tree V and VI, and receiver operating characteristic (ROC) curves were generated. As shown in Figure 5, combining the markers from each tree with log-transformed CEA values yielded a higher area under the ROC curve than for either alone.
In this study, five biomarker proteins were detected that were able to reliably distinguish colorectal cancer patients and healthy controls using the SELDI-TOF MS technique for protein profiling. Two of these were identified as apolipoprotein C-I (6.6×103 Da) and apolipoprotein A-I (28×103 Da). Using the ProteinChip Software, the 3.3×103-Da protein could be identified as a doubly charged form of the 6.6×103-Da apolipoprotein C-I, which was confirmed by its appearance in the MS spectrum of pure apolipoprotein C-I isolated from a gel. The m/z of 3.1×103 was found to be an N-terminal fragment of albumin. In addition, the detection of these biomarkers’ expression difference was shown to be reproducible on two separate occasions, considering the obtained classification tree sensitivity and specificity between 65% and 90% when using the second sample set as a blinded validation set. Such reproducible detection is imperative for any future use as a clinical tool. Sensitivities and specificities obtained with data from the blinded sample set were comparable to those obtained by cross-validation within one sample set (data not shown), which supports the fact that there was no additional misclassification of samples due to experimental variability between the two sample sets.
Several reports have been made of differential expression of the same m/z values in colorectal cancer, even though different chip surfaces were used. Yu et al[17] reported a 3 329- and 6 669-Da protein to be differentially expressed on a hydrophobic chip surface, that were not selected in the final diagnostic pattern. In the same study, a 4 477-Da protein, which was a classifier in the final pattern, was also up-regulated in colorectal cancer patients[17,18]. The 6.6×103- and 3.3×103-Da proteins were detected by Yu et al[17] but not identified. It is likely that these are apolipoprotein C-I and its doubly charged form, since this is a very hydrophobic protein and retention on a hydrophobic chip surface is very plausible. In fact, in our study population, these m/z values were also seen on hydrophobic chips (data not shown). This appearance of (likely) the same colorectal cancer biomarkers in different laboratories underlines their validity. Also, a 5.9×103-Da protein was reported, which was an up-regulated biomarker in serum of colorectal cancer patients[17,18]. Despite lack of significance in our study, there was a tendency for our 5.9×103-Da protein to be higher in patients than controls, which is consistent with the result from Yu et al[17] and indicates this may be the same protein. Data from a proteomic study by our group[19] on breast cancer patients have shown a 5.9×103-Da down-regulated protein in this cancer type, which is in concordance with the 5.9×103-Da protein in the current study. The protein in the former study was identified as a fragment of fibrinogen alphaE chain.
Apolipoprotein C-I is primarily synthesised in the liver and, to a minor degree, in the small intestine. Its function resides mainly in lipoprotein metabolism[20]. It is originally formed as a pro-peptide of 9.3×103 Da, which generates the mature protein upon cleavage during translation. To our knowledge, no previous reports about apolipoprotein C-I down-regulation in cancer have been made as yet. However, a 6.6×103-Da protein was detected and identified as apolipoprotein C-I in another SELDI-TOF MS study, being decreased in hemorrhagic versus ischemic stroke and hemorrhagic stroke versus controls on a strong anion exchange surface at pH 9[21]. Apolipoprotein A-I is synthesised both in the liver and small intestine and is a major constituent of HDL apolipoprotein. It is a known negative acute phase reactant, of which decreased expression has been described in several cancers, including a SELDI-TOF MS study on ovarian cancer[22-25]. In the latter study an immunoassay was peformed. In contrast to our data, the authors found no decreased apolipoprotein A-I levels in colorectal cancer. However, apolipoprotein A-I levels assessed by immunoassay may reflect concentrations of both bound pro-apolipoprotein A-I and apolipoprotein A-I. Increased expression of apolipoprotein A-I has been described in tissue of both liver metastases and, to a lesser extent, primary tumours of colorectal origin[26]. The observed decrease in serum levels in our study thus may be due to decreased liver synthesis. Other human proteomics studies in which differential expression of apolipoprotein A-I has been described include a SELDI-TOF MS analysis of plasma from patients with diabetes, and several studies using 2D-gel electrophoresis in old versus young brain tissue, cerebrospinal fluid of patients with Alzheimer’s disease, serum during infection with hepatitis B virus, and plasma during acute coronary syndrome[27-31]. In all these diseases, decreased levels of apolipoprotein A-I were observed. To our knowledge, the albumin fragment that was found in this study has not been described in the literature before. Albumin is synthesised with an 18-amino acid signal peptide and a 6-amino acid pro-peptide. Over-expression of this specific fragment may be caused by enhanced proteolytic activity, as increased proteolysis is common in cancer invasion and metastasis[32]. However, for this fragment, a correlation with sample age was seen in the colorectal cancer group (data not shown). Thus, it cannot be ruled out that it is a product of protein degradation upon storage.
Although the identification of apolipoprotein C-I and apolipoprotein A-I as biomarkers suggests an acute phase response, comparison with routine markers for establishing such a response, CRP and TRF, shows that our biomarkers are much more sensitive for colorectal cancer than these. The value of our biomarkers for detection of colorectal cancer was also evaluated by comparison with the predictive value of CEA. Sensitivity of our biomarkers was higher than that of CEA considering all samples. Stratification by Dukes’ stages showed a significant better sensitivity of our classification trees (91.7%, 11/12) compared to CEA (25.0%, 3/12) in Dukes’ C colorectal cancer, although at stage D CEA performed better. Combining log-transformed CEA in a logistic regression model with the markers in the trees resulted in a higher AUC in the ROC curve than for either log(CEA) or the combined tree classifiers alone. This indicates that our markers provide additional information to CEA values. CEA sensitivity has been reported to be lower in earlier stages of colorectal cancer. Sensitivity has been reported to vary between 3% and 66.7% for Dukes’ A to D staged disease[2,18,33]. No conclusions can be drawn on the performance of our classification trees at earlier stages of colorectal cancer due to limited samples, but 2 of 3 patient samples from stage A and B were correctly classified by the trees and none when using the clinical cut-off for CEA.
Our results showed that it is very important to compare any biomarkers found for a certain type of cancer with those for other tumour types. This is lacking in most of the SELDI-TOF MS studies published so far. We found that most of our biomarker proteins were differentially expressed in other cancers as well. Lack of a significant difference at m/z 3.3×103 and 4.5×103 in breast cancer patients could be explained by the large proportion of early disease (9 of 10 stage 2) in this group compared to the others (mainly stage 3 and 4). However, since m/z 3.3×103 is the doubly charged 6.6×103-Da protein, which was significantly less expressed in breast cancer, this lack of significance is more likely due to slight differences in ionisation of this protein in this group. The fact that the 3.1×103-Da protein is not significantly different in ovarian and prostate cancer may result from the limited sample size, as for the smaller colorectal cancer group in this analysis with comparable mean peak intensity no significant difference at m/z 3.1×103 was observed either, although in sample sets A and B it was.
Even though most of our biomarkers are not specific for colorectal cancer, a potential role for them lies in therapy evaluation, disease surveillance or prognosis, possibly combined with CEA or other available markers. At present, CEA is recommended for monitoring chemotherapy, but no studies showing any benefit on survival, quality of life, or reduction of costs are available, although serial CEA testing may lead to earlier detection of progressive disease[5]. In addition, specificity of CEA for treatment monitoring can be compromised by transient increases during treatment with various chemotherapeutic drugs, such as 5-flourouracil and levamisole[5]. Since the expression profiles of our reported markers reliably reflect presence of cancer, be it colorectal cancer or not, changes in expression levels may correspond to response to therapy or disease progression and provide additional information to CEA levels.
In conclusion, our results show that SELDI-TOF MS is a suitable technique to find new serum biomarkers for colorectal cancer. The markers we have found in this study reliably distinguish colorectal cancer patients from healthy persons. Although not specific for colorectal cancer, they have a potential role as markers in treatment monitoring, disease surveillance, or prognosis, possibly in combination with other available markers. To extend the study population and evaluate the ability of our biomarkers to detect early-stage tumours and polyps, a prospective study is currently ongoing.
| 1. | Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704-2714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 529] [Article Influence: 24.0] [Reference Citation Analysis (3)] |
| 2. | Duffy MJ. Carcinoembryonic antigen as a marker for colorectal cancer: is it clinically useful? Clin Chem. 2001;47:624-630. [PubMed] |
| 3. | Ahlquist DA, Wieand HS, Moertel CG, McGill DB, Loprinzi CL, O'Connell MJ, Mailliard JA, Gerstner JB, Pandya K, Ellefson RD. Accuracy of fecal occult blood screening for colorectal neoplasia. A prospective study using Hemoccult and HemoQuant tests. JAMA. 1993;269:1262-1267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 126] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 4. | Greenberg PD, Bertario L, Gnauck R, Kronborg O, Hardcastle JD, Epstein MS, Sadowski D, Sudduth R, Zuckerman GR, Rockey DC. A prospective multicenter evaluation of new fecal occult blood tests in patients undergoing colonoscopy. Am J Gastroenterol. 2000;95:1331-1338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 64] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Duffy MJ, van Dalen A, Haglund C, Hansson L, Klapdor R, Lamerz R, Nilsson O, Sturgeon C, Topolcan O. Clinical utility of biochemical markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines. Eur J Cancer. 2003;39:718-727. [RCA] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 254] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 6. | Bast RC, Ravdin P, Hayes DF, Bates S, Fritsche H, Jessup JM, Kemeny N, Locker GY, Mennel RG, Somerfield MR. 2000 update of recommendations for the use of tumor markers in breast and colorectal cancer: clinical practice guidelines of the American Society of Clinical Oncology. J Clin Oncol. 2001;19:1865-1878. [PubMed] |
| 7. | Issaq HJ, Veenstra TD, Conrads TP, Felschow D. The SELDI-TOF MS approach to proteomics: protein profiling and biomarker identification. Biochem Biophys Res Commun. 2002;292:587-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 374] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 8. | Graves PR, Haystead TA. Molecular biologist's guide to proteomics. Microbiol Mol Biol Rev. 2002;66:39-63; table of contents. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 367] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 9. | Adam BL, Qu Y, Davis JW, Ward MD, Clements MA, Cazares LH, Semmes OJ, Schellhammer PF, Yasui Y, Feng Z. Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 2002;62:3609-3614. [PubMed] |
| 10. | Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572-577. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2358] [Cited by in RCA: 1925] [Article Influence: 80.2] [Reference Citation Analysis (0)] |
| 11. | Zhukov TA, Johanson RA, Cantor AB, Clark RA, Tockman MS. Discovery of distinct protein profiles specific for lung tumors and pre-malignant lung lesions by SELDI mass spectrometry. Lung Cancer. 2003;40:267-279. [RCA] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 128] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 12. | Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48:1296-1304. [PubMed] |
| 13. | Shiwa M, Nishimura Y, Wakatabe R, Fukawa A, Arikuni H, Ota H, Kato Y, Yamori T. Rapid discovery and identification of a tissue-specific tumor biomarker from 39 human cancer cell lines using the SELDI ProteinChip platform. Biochem Biophys Res Commun. 2003;309:18-25. [RCA] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 82] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 14. | Krieg RC, Fogt F, Braunschweig T, Herrmann PC, Wollscheidt V, Wellmann A. ProteinChip Array analysis of microdissected colorectal carcinoma and associated tumor stroma shows specific protein bands in the 3.4 to 3.6 kDa range. Anticancer Res. 2004;24:1791-1796. [PubMed] |
| 15. | Wang YY, Zhang Z, White N, Rosenzweig J, Li J, Shih I, Sokoll LJ, Chan DW. Detection of cancer biomarkers by SELDI proteomics technology from serum in colorectal carcinoma. Proc Amer Assoc Cancer Res. 2003;44. |
| 16. | Zhao G, Gao CF, Song GY, Li DH, Wang XL. [Identification of colorectal cancer using proteomic patterns in serum]. Ai Zheng. 2004;23:614-618. [PubMed] |
| 17. | Yu JK, Chen YD, Zheng S. An integrated approach to the detection of colorectal cancer utilizing proteomics and bioinformatics. World J Gastroenterol. 2004;10:3127-3131. [PubMed] |
| 18. | Chen YD, Zheng S, Yu JK, Hu X. Artificial neural networks analysis of surface-enhanced laser desorption/ionization mass spectra of serum protein pattern distinguishes colorectal cancer from healthy population. Clin Cancer Res. 2004;10:8380-8385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 108] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 19. | Gast MCW, Bonfrer JMG, Rutgers E, Schellens JHM, Beijnen JH. Proteomics in patients with breast cancer: unique profile discriminates patients from matched controls: Proceedings of the Dutch Society for Clinical Pharmacology and Biopharmacy, 16 April 2004. Br J Clin Pharmacol. 2005;59:123-139. [RCA] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 20. | Jong MC, Hofker MH, Havekes LM. Role of ApoCs in lipoprotein metabolism: functional differences between ApoC1, ApoC2, and ApoC3. Arterioscler Thromb Vasc Biol. 1999;19:472-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 407] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 21. | Allard L, Lescuyer P, Burgess J, Leung KY, Ward M, Walter N, Burkhard PR, Corthals G, Hochstrasser DF, Sanchez JC. ApoC-I and ApoC-III as potential plasmatic markers to distinguish between ischemic and hemorrhagic stroke. Proteomics. 2004;4:2242-2251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 95] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 22. | Steel LF, Shumpert D, Trotter M, Seeholzer SH, Evans AA, London WT, Dwek R, Block TM. A strategy for the comparative analysis of serum proteomes for the discovery of biomarkers for hepatocellular carcinoma. Proteomics. 2003;3:601-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 119] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 23. | Ryu JW, Kim HJ, Lee YS, Myong NH, Hwang CH, Lee GS, Yom HC. The proteomics approach to find biomarkers in gastric cancer. J Korean Med Sci. 2003;18:505-509. [PubMed] |
| 24. | Wang Z, Yip C, Ying Y, Wang J, Meng XY, Lomas L, Yip TT, Fung ET. Mass spectrometric analysis of protein markers for ovarian cancer. Clin Chem. 2004;50:1939-1942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 25. | Zhang Z, Bast RC, Yu Y, Li J, Sokoll LJ, Rai AJ, Rosenzweig JM, Cameron B, Wang YY, Meng XY. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64:5882-5890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 703] [Cited by in RCA: 683] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 26. | Tachibana M, Ohkura Y, Kobayashi Y, Sakamoto H, Tanaka Y, Watanabe J, Amikura K, Nishimura Y, Akagi K. Expression of apolipoprotein A1 in colonic adenocarcinoma. Anticancer Res. 2003;23:4161-4167. [PubMed] |
| 27. | He QY, Lau GK, Zhou Y, Yuen ST, Lin MC, Kung HF, Chiu JF. Serum biomarkers of hepatitis B virus infected liver inflammation: a proteomic study. Proteomics. 2003;3:666-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 125] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 28. | Puchades M, Hansson SF, Nilsson CL, Andreasen N, Blennow K, Davidsson P. Proteomic studies of potential cerebrospinal fluid protein markers for Alzheimer's disease. Brain Res Mol Brain Res. 2003;118:140-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 227] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 29. | Dayal B, Ertel NH. ProteinChip technology: a new and facile method for the identification and measurement of high-density lipoproteins apoA-I and apoA-II and their glycosylated products in patients with diabetes and cardiovascular disease. J Proteome Res. 2002;1:375-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 30. | Mateos-Cáceres PJ, García-Méndez A, López Farré A, Macaya C, Núñez A, Gómez J, Alonso-Orgaz S, Carrasco C, Burgos ME, de Andrés R. Proteomic analysis of plasma from patients during an acute coronary syndrome. J Am Coll Cardiol. 2004;44:1578-1583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 99] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 31. | Chen W, Ji J, Xu X, He S, Ru B. Proteomic comparison between human young and old brains by two-dimensional gel electrophoresis and identification of proteins. Int J Dev Neurosci. 2003;21:209-216. [RCA] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 32. | Garbett EA, Reed MW, Brown NJ. Proteolysis in colorectal cancer. Mol Pathol. 1999;52:140-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 33. | Kim SB, Fernandes LC, Saad SS, Matos D. Assessment of the value of preoperative serum levels of CA 242 and CEA in the staging and postoperative survival of colorectal adenocarcinoma patients. Int J Biol Markers. 2003;18:182-187. [PubMed] |
S- Editor Wang J L- Editor Kumar M E- Editor Bi L