Published online Feb 28, 2005. doi: 10.3748/wjg.v11.i8.1131
Revised: August 3, 2004
Accepted: September 19, 2004
Published online: February 28, 2005
AIM: To study the role of intestinal flora in inflammatory bowel disease (IBD).
METHODS: The spatial organization of intestinal flora was investigated in normal mice and in two models of murine colitis using fluorescence in situ hybridization.
RESULTS: The murine small intestine was nearly bacteria-free. The normal colonic flora was organized in three distinct compartments (crypt, interlaced, and fecal), each with different bacterial compositions. Crypt bacteria were present in the cecum and proximal colon. The fecal compartment was composed of homogeneously mixed bacterial groups that directly contacted the colonic wall in the cecum but were separated from the proximal colonic wall by a dense interlaced layer. Beginning in the middle colon, a mucus gap of growing thickness physically separated all intestinal bacteria from contact with the epithelium. Colonic inflammation was accompanied with a depletion of bacteria within the fecal compartment, a reduced surface area in which feces had direct contact with the colonic wall, increased thickness and spread of the mucus gap, and massive increases of bacterial concentrations in the crypt and interlaced compartments. Adhesive and infiltrative bacteria were observed in inflamed colon only, with dominant Bacteroides species.
CONCLUSION: The proximal and distal colons are functionally different organs with respect to the intestinal flora, representing a bioreactor and a segregation device. The highly organized structure of the colonic flora, its specific arrangement in different colonic segments, and its specialized response to inflammatory stimuli indicate that the intestinal flora is an innate part of host immunity that is under complex control.
-
Citation: Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: A fluorescence
in situ hybridization study in mice. World J Gastroenterol 2005; 11(8): 1131-1140 - URL: https://www.wjgnet.com/1007-9327/full/v11/i8/1131.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i8.1131
The general consensus that the intestinal flora is important in the pathogenesis of chronic bowel inflammation is based on solid clinical and experimental evidence[1]. Despite this, all attempts to identify specific changes of the intestinal flora that are associated with chronic inflammation have either failed or are inconsistent[2]. Previous investigations of bacterial involvement in inflammatory bowel disease (IBD) have mainly been based on comparative studies of bacterial isolates. It is likely however, that the intestinal flora is structurally organized[3]. Sampling of the intestinal contents prior to culture or gene-based identification disrupts the structural organization of bacteria within the gut and may disguise the complex interactions between intestinal flora and the host.
The aim of this work was to study the composition and spatial organization of the intestinal flora in normal mice and in two commonly used models of murine colonic inflammation, using fluorescence in situ hybridization (FISH) with rRNA-targeted, fluorescent Cy3/Cy5 (carbocyanine) labeled oligonucleotide probes[4].
Three different groups of mice were investigated: wild-type mice without colitis (WT group, n = 3), wild-type mice with acute chemically-induced colitis (DSS group, n = 3), IL-10 knockout mice without colitis (IL-10 C- group, n = 2) and IL-10 knockout mice with manifest colitis (IL-10 C+ group, n = 3). All mice studied were of C57BL/6J background and obtained from Jackson Laboratories (Bar Harbor, ME). Mice were housed in micro-isolator racks and allowed access to food and water ad libitum. Colitis was induced in 5 to 8-wk-old wild type mice (WT) by adding 3% dextran sulfate sodium (DSS, 40 kDa MW; obtained from ICN, Costa Mesa, CA) to the drinking water for 5 d. Acute colitis developed in all treated animals by d 4 of DSS exposure. Symptoms included diarrhea, gross or occult blood in stool (measured by Hemoccult II Sensa cards; Beckman Coulter, Palo Alto, CA), and weight loss. DSS-treated mice were sacrificed on d 7 after first exposure to DSS. The spontaneous development of colitis in mice genetically deficient in IL-10 was monitored by daily weight measurements and weekly stool occult blood tests. Animals were sacrificed by CO2 asphyxiation. A portion of the midjejunum, the terminal ileum and the whole colon were obtained. The colon was divided into 5 parts representing the cecum, proximal, mid, distal colon, and the rectum. Each colon part was analyzed separately. Intestinal segments were fixed in Carnoy’s solution[5] for 3 h, then processed into paraffin blocks using standard techniques, cut into 4-10 µm sections and placed on SuperFrost slides (R. Langenbrinck, Emmendingen, Germany) for pathologic examination and FISH studies.
The studies were conducted under protocols approved by the Institutional Animal Care and Use Committee of Duke University Medical Center.
Oligonucleotide probes were synthesized with a Cy3 or Cy5 reactive fluorescent dye at the 5’ end (MWG Biotech, Ebersberg, Germany). Forty domain, group and species-specific FISH probes (Table 1) were applied to murine colon sections. The Eub338 universal bacterial probe was used to detect virtually all relevant bacteria. The nonsense probe Non338 was used to test for nonspecific binding of oligonucleotide probes. Formamide concentration and hybridization temperature were used as described to achieve the optimal stringency[6-30]. Additional hybridizations using a permeation step with lysozyme were performed in parallel for detection of Gram-positive bacteria.
| Name | Target | Reference |
| Eub338 | Virtually all Bacteria, Kingdom Eubacteria | 6 |
| Alf1b | Alpha group of Proteobacteria: Rhodobacter, Acetobacter, Paracoccus | 7 |
| Beta42a | Beta subclass of Proteobacteria: Rhodocyclus, Bordetella, Neisseria,Thiobacillus, Alcaligenes and other | 7 |
| Gam42a | Gamma subclass of Proteobacteria: Enterobacteriaceae, Proteus, Legionella, Azotobacter | 7 |
| Ebac | Enterobacteriaceae | 8 |
| Ec1531 | Escherichia coli | 9 |
| Y16s-69 | Yersinia species | 10 |
| Srb385 | Sulfate reducing bacteria | 11 |
| Sgd | Desulfotomaculum subgroup | 12 |
| Hpy-1 | Helicobacter pylori epsilon subclass of Proteobacteria | 13 |
| Arc1430 | Arcobacter ssp. epsilon subclass of Proteobacteria | 14 |
| HGC | Gram-positive bacteria with high G+C content:Actinobacteria | 15 |
| LGC | Gram-positive bacteria with low G+C content: Firmicutes | 16 |
| Sfb | Segmented filamentous bacteria | 17 |
| Erec | Clostridium coccoides - Eubacterium rectale group | 18 |
| Lach | Subgroup of Erec | 19 |
| Ehal | Subgroup of Erec | 19 |
| Chis150 | Clostridium histolyticum group (Incl. Clostridiumperfringens and Clostridium botulinum) | 18 |
| Clit135 | Clostridium lituseburense group (incl. Clostridium difficile) | 18 |
| Lab158 | Lactobacillus and Enterococcus group | 20 |
| Strc493 | Streptococcus group | 18 |
| Enc131 | Enterococcus spp and other | 21 |
| Efaec | Enterococcus faecalis, Enterococcus sulfuricus | 22 |
| Ato291 | Atopobium, Coriobacterium, Eggerthella and Collinsella spp | 23 |
| Cor653 | Coriobacterium group | 23 |
| Ecyl | Eubacterium cylindroides, Clostridium innocuum, Eubacterium biforme, Eubacterium tortuosum and other | 19 |
| Phasco | Phascolarctobacterium faecium, Acidaminococcus fermentans, and Succiniclasticum ruminis | 19 |
| Veil | Veillonella group | 19 |
| Rbro, Rfla | Clostridium sporosphaeroides, Ruminococcus bromii, andClostridium leptum, Ruminococcus albus and Ruminococcus flavefaciens | 19 |
| UroA, UroB | Ruminococcus obeum-like bacteria (subgroup of Erec) | 24 |
| Ser1410 | Genus Brachyspira | 25 |
| Bif164 | Bifidobacterium | 26 |
| CF319a | Cytophaga-Flavobacteria group | 27 |
| Bac303 | Bacteroides/Prevotella group | 27 |
| Bfra602 | Bacteroides fragilis group | 18 |
| Bdis656 | Bacteroides distasonis group | 18 |
| Fprau | Fusobacterium prausnitzii group | 28 |
| Dss658 | Sulfate reducing bacteria subgroup of delta proteobacteria | 29 |
| Arch915 | Archaea | 30 |
| Non338 | Nonsense probe used to test for nonspecific binding | 7 |
In situ quantification of fecal bacteria Bacteria were visualized by FISH and 4, 6-diamidino-2-phenylindole stain (DAPI) under a Nikon e600 fluorescence microscope (Nikon, Tokyo, Japan) and photo-documented with a Nikon DXM1200 camera and software (Nikon). The enumeration of bacteria was performed only when hybridization signals were clear and morphologically distinguishable as bacterial cells by triple identification with universal and group-specific FISH probes and DAPI stain, in absence of cross-hybridization or hybridization with the Non338 nonsense probe.
For each group-specific FISH probe, high power (×1000 magnification) photographs were made of three different microscopic fields within the intestinal lumen at the widest, narrowest, and median diameters of each colonic segment. The estimation of bacterial concentrations was made based on the assumption that a 10-µL sample with a cell concentration of 107 cells per mL brings 40 cells per average microscopic field at magnification of 1000[4]. Two investigators independently counted bacteria within a 50 µm2 area of the microscopic field (about 1 cm2 of the 100% scale photographic image). The mean number of bacteria was recorded for each colonic segment. In cases where single bacterial cells were morphologically indistinguishable due to high bacterial concentrations and the fluorescence was confluent to a homogeneously fluorescent mass, no enumeration was attempted and the bacterial concentration was assigned a value of >1012 per mL. If single bacterial cells were not distinguishable within the bacterial carpet but some empty gaps could be recognized, the bacterial concentration was assigned a value of >1011 per mL.
Evaluation of spatial interrelationship of bacteria and assessment of cross-hybridization The spatial organization of bacteria was evaluated in three steps. Group or species-specific signals (green-orange fluorescence Cy3) were visualized simultaneously with a Eub338 probe universal for all relevant bacteria (dark red fluorescence Cy5). All probes positively hybridizing with more than 1% of bacteria were further combined with each other in pairs, in a two-color analysis (Cy3 vs Cy5). This allowed us to characterize the position of different bacterial groups relative to each other within the intestine and to assess potential cross-hybridization. Although probes chosen for this study were designed for definite bacterial groups and extensively tested (Table 1)[6-30], the specificity of each probe is relative. In these experiments, multiple observations were made to determine whether bacteria that hybridized with one probe gave a positive hybridization signal with a probe representing other bacterial groups, rather than to rely on results obtained with a single probe. When probes of unrelated bacterial groups hybridized with the same bacteria, the hybridization stringency was adjusted until a clear differentiation was possible. Otherwise the results achieved with cross-hybridizing probes were excluded.
Quantification of dynamic compartments The quantification of the dynamic structures, such as bacterial populations of crypts, bacteria organized in layers adjacent to the mucosa, and the width and spread of the mucus, was based on the mean values of at least ten photographs of different microscopic fields made within each bowel segment at a magnification of ×400 for layers and ×1000 for crypts.
Mean values and standard deviations were calculated for bacteria concentrations and measurements of layer (e.g., mucus or interlaced) thickness. Using t-tests, P<0.05 was considered statistically significant.
Healthy wild type mice The ileum was narrow. The few microorganisms found were heterogeneously composed, random, and without signs of adhesion or contact with the intestinal wall (Figure 1).
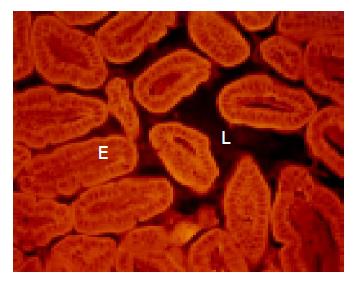
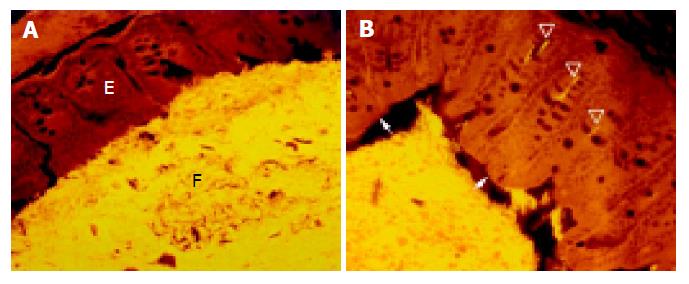
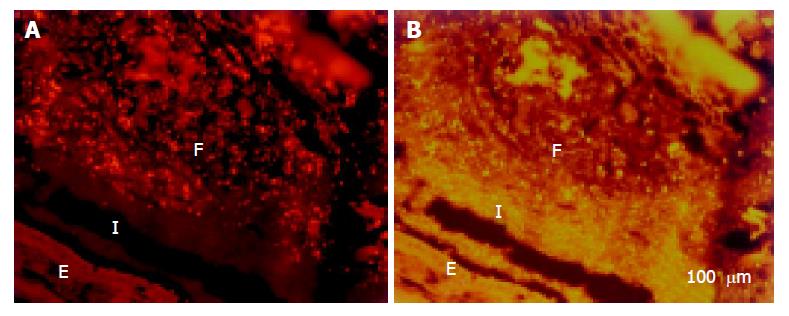
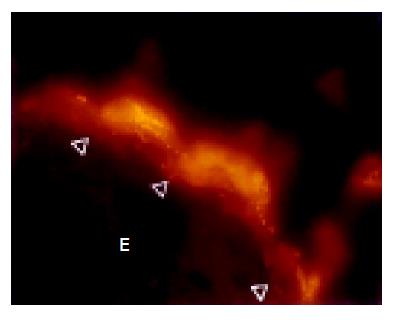
The cecum was wide and contained a highly concentrated mass of bacteria. The exact enumeration of bacteria was impossible since single bacteria could not be distinguished within the confluent fluorescent carpet (Figure 2A). Luminal bacteria were in direct contact with the wall of the cecum. Despite this contact, the bacteria lining the cecal mucosa were probably non-adherent. Shrinkage caused by fixation sometimes led to dissection of feces from the cecal wall. In all regions where this dissection took place, no bacteria adhered to the mucosal surface (Figure 2B). Abundant bacteria were observed within crypts (Figure 2B, arrowheads).
The proximal colon was wide initially, and narrows distally. Fecal bacteria were present in high numbers and contacted the colonic wall along most of its length. Numerous bacteria were seen in nearly half of all intestinal crypts, even very deep within the wall. The number of crypts containing bacteria and the number of bacteria within these crypts were even higher in the proximal colon than in the cecum.
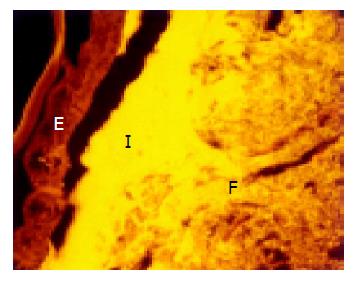
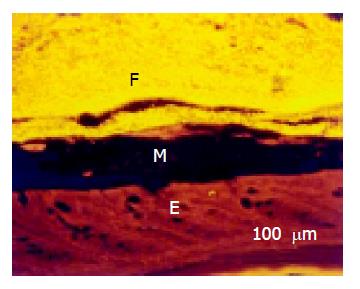
A thin but dense band of homogeneous bacteria was present in the distal part of the colon. Because this band of bacteria was interlaced between the epithelial wall and the unorganized fecal masses, it was termed the interlaced layer (Figure 3).
The mid-colon was narrow, and generally contained little feces. If present, fecal material was restricted to a fine tube-like structure located centrally within the intestinal lumen and separated from the epithelial surface by mucus. Bacteria were seen less often within crypts (Figure 4).
The colon lumen widened in the distal colon and was filled with masses of feces containing a high concentration of bacteria. A growing mucus gap devoid of bacteria completely separated the colonic wall from this fecal biomass. Nearly no bacteria was found in crypts and no interlaced layer was seen.
The lumen of rectum was wide. The mucus layer separating the bacteria from the mucosa continuously thickened (Figure 5). No bacteria were found within crypts in the rectum.
IL-10 knockout mice without colitis The bacterial concentrations in the ileum of IL-10 knockout mice without colitis were similar to findings in healthy WT mice. However, the concentrations and occurrence of bacteria in crypts, the thickness of the interlaced layer, and the overall concentrations of fecal bacteria were noticeably less compared to healthy WT mice (Table 2).
| Group | Ileum | Cecum | Proximal | Middle | Distal | Rectum | |
| Mean lumen width in | WT | Narrow 0.2 | Broad 6.4 | Narrowing 2.9 | Narrow <0.1 | Widening 2.4 | Broad 2.5 |
| microscopic fields | DSS | Narrow 0.1 | Broad 3.0 | Irregular 0.9 | Irregular 0.8 | Irregular 0.6 | Irregular 0.6 |
| (×1000) between | IL-10 C- | Narrow 0.1 | Broad 2.1 | Irregular 0.6 | Irregular 0.5 | Irregular 0.7 | Irregular 1.2 |
| opposite walls | IL-10 C+ | Narrow 0.3 | Broad 2.0 | Irregular 0.6 | Irregular 0.5 | Irregular 0.5 | Narrow 0.2 |
| Min -max | WT | 0.2-2.5×107 | >1012 | >1012 | - | >1012 | >1012 |
| concentrations of | DSS | <107 | 1.3-6.7×109 | 1.2-4.0×109 | 2.1-6.6×109 | 1.8-2.2×109 | 1.1-2.2×109 |
| bacteria per mL | IL-10 C- | <107 | 1.0-5.3×1010 | 1.0-5.2×1010 | 0.6-3.4×1010 | 1.2-1.9×1010 | 1.4-2.3×1010 |
| of feces | IL-10 C+ | <107 | 1.7-5.0×109 | 0.7-4.2×109 | 0.7-4.2×109 | 4.7-7.1×109 | 4.7-9.2×109 |
| Min -max. length | WT | None | None | 8-10% | 80-90% | Complete | Complete |
| of mucus gap | DSS | None | 8-20% | 40-50% | Complete | Complete | Exudate |
| separating feces | IL-10 C- | None | 5-10% | 70-80% | Complete | Complete | Complete |
| from epithelium | IL-10 C+ | None | 10-20% | 50-80% | Complete | Complete | Exudate |
| Min-max width | WT | - | 0 | 0- 5 | 0-20 | 20-50 | 30-100 |
| of the mucus in | DSS | - | 0- 5 | 0-10 | 10-50 | 40-75 | 60-150 |
| µm between | IL-10 C- | - | 0-10 | 0-10 | 15-50 | 40-100 | 60-200 |
| epithelium and feces | IL-10 C+ | - | 0-20 | 0-20 | 5-50 | 60-200 | Infiltrate |
| Mean percent of | WT | - | 10% (3-12) | 16% (6-16) | 5% (single) | <1% (single) | 0 |
| crypts with bacteria | DSS | - | 22% (4-25) | 44% (9-30) | 27% (4-15) | 16% (2-9) | <1% |
| (mean-max number of | IL-10 C- | - | <1% (single) | 5% (1-3) | 2% (single) | 1% (single) | No |
| bacteria within each) | IL-10 C+ | 28% ( 3-20) | 34% (8-34) | 27% (2-20) | 16% (3-12) | <1% | |
| Min-max thickness | WT | - | 0 | 5-25 | 0-10 | 0 | 0 |
| of the interlaced | DSS | - | 0-100 | 80-400 | 8-40 | 0-2 | 0-2 |
| layer (µm) | IL-10 C- | - | 2-10 | <5 | 0 | 0-2 | 0 |
| IL-10 C+ | 0-120 | 200-500 | 80-150 | 0-15 | Infiltrate |
DSS-treated wild-type mice and IL-10 knockout mice with colitis Five striking changes were observed both in mice with colitis due to DSS exposure and in IL-10 knockout mice with spontaneous severe colitis (Table 2). First, there was a massive reduction of bacterial concentrations within the fecal masses in all colonic segments compared to healthy WT mice. Second, in inflamed colon, the middle colon lost its dividing function. The narrowing observed in normal WT mice was not observed in the colitis groups. The area where fecal masses had direct contact with the mucosal surface was reduced. The mucus gap separating the colonic wall from the fecal biomass began more proximally than in healthy WT mice. In mice with colitis, the mucus layer was evident in the distal cecum and became broad and complete in the proximal colon. Third, the number and occurrence of bacteria within crypts increased manifold compared to mice without colitis. Bacteria could be regularly seen even in crypts of the inflamed distal colon, while bacteria were seen only in the crypts of cecum and proximal colon in healthy WT mice. Fourth, the interlaced layer became extremely thick in the proximal and middle colon of mice with colitis, and reached thicknesses of >300 µm in some locations. The interlaced layer could be observed separating feces from mucosa, beginning in the distal cecum and continuing to the proximal portion of the distal colon. Fifth, in addition to these gradual changes, bacteria adhering to the mucosa or invading mucosal epithelial cells were seen in mice with colitis, but not in healthy WT mice.
Intestinal bacteria were organized in crypt, interlaced, fecal, adhesive, and invasive compartments. Crypt, interlaced, and fecal compartments were observed in all groups; however, adhesive and invasive bacteria were seen only in animals with colitis. The bacterial groups found in different compartments are summarized in Table 3. Generally all bacterial groups that were observed within the crypt compartment could also be found in the interlaced and fecal compartments. However, not all bacterial groups found in feces had access to the interlaced compartment. In addition, some bacterial groups seen in both fecal and interlaced compartments were never seen in the crypt compartment (Table 3).
The composition of the bacterial populations within single compartments (feces, interlaced, crypt) was the same in all investigated groups of animals. However, the spread, extent, and volume of these different compartments within different colonic segments differed markedly between groups with and without colitis, irrespective of the etiology of the inflammation.
Fecal compartment The composition of the bacterial community in the lumen of ileum could not be reliably evaluated in all groups of mice due to low bacterial concentrations and their irregular distribution within the lumen. Different bacterial groups found in the ileum appeared to be at random and varied from animal to animal in an unpredictable manner.
All bacterial groups that were positively hybridized with FISH probes were homogeneously mixed within the feces, without a gradient in distribution between regions adjacent to the mucosa and distant regions (Figure 6). The Arch915, Gam42a, Beta42a, Srb385, Bfra602, CF319a probes demonstrated a high grade of cross-hybridization and were excluded from the evaluation. The Erec, Lach, Alf1b, Phasco, Lab158 and Bac303 probes hybridized with more than 10% of the fecal population at least in one of the animals tested. The Rbro, Chis150, Clit135, Ehal, Ecyl, LGC, Cor653, Bdis659 probes hybridized with more than 1% of the fecal population. The Bif164, HGC, Rfla, Enc131, Veil, Ato291, UroB, UroA, Strc493, Ebac, Arc1430 probes hybridized with less than 1% of the population. Despite this low overall proportion, the absolute number of these bacterial groups was higher than 107 cells/mL and therefore, easy to distinguish from the non-specific background. The Ec1531, Y16s-69, Sgd, Fprau, Sfb, Hpy-1, Efaec, Ser1410, Dss658 probes failed to give signals that were different from the background fluorescence seen with the nonsense probe.
Changes in bacterial concentrations within the fecal compartment from the cecum to the rectum The types of bacteria present in the fecal compartment were constant in all colonic segments and animal groups; however, the concentrations differed markedly. This was illustrated by the changes in mean concentrations of bacteria that hybridized with Erec and Bac303 probes, the two most abundant bacterial groups within the fecal compartment, during the transition from cecum to rectum (Table 4).
| Probe | Group | Cecum | Rectum |
| Erec | WT | 80×109 | 0.8×109 |
| DSS | 13×108 | 0.3×108 | |
| IL-10 C- | 15×108 | 25×108 | |
| IL-10 C+ | 8×108 | 12×108 | |
| Bac | WT | 2.8×108 | 2.1×108 |
| DSS | 1×108 | 0.5×108 | |
| IL-10 C- | 5×108 | 2×108 | |
| IL-10 C+ 28 wk | 11×108 | 5×108 |
Bacterial groups found both in the crypt and fecal compartments (e.g., Erec, Alpf1b, Phasco, etc.; Table 3) were predominant in the cecum of healthy WT mice and their concentrations declined distally. The concentrations of bacterial groups found mainly in feces (e.g., Bacteroides, Clit135, Chis150, etc. Table 3) remained relatively stable throughout the colon. Thus their concentrations increased relative to the Erec-like groups.
Crypt bacterial communities Crypt bacteria were mainly found in the cecum and proximal colon of healthy WT mice (Table 2). Starting with the middle colon, bacteria could be only sporadically observed within the crypts. No bacteria were found within crypts of the distal colon and rectum. The occurrence and number of crypt bacteria were significantly reduced in IL-10 knockout mice without colitis. In contrast, in both DSS mice with colitis and IL-10 knockout mice with spontaneous colitis, the crypt population was amplified and crypt bacteria could be seen sporadically even in the rectum (Table 2). Despite this significant difference in the extent and the distribution of crypt communities, the composition of the crypt population was the same in all investigated animal groups. Groups hybridizing with the Alf1b, Erec (Lach), Phasco and Lab158 probes were detected within crypts in different combinations. In the cecum, crypt bacteria directly contacted fecal masses and these bacteria were the main constituents of the fecal compartment. Bacteria positively hybridizing with Bac303 (Bacteroides), Chis150 (Clostridium histolyticum), and Clit135 (including Clostridium difficile) probes were never identified within crypts of any of the groups investigated (Table 3), although they were homogeneously intermixed within the fecal compartment and directly contacted the mucosa and crypt mouths in the cecum. In the proximal colon, the groups of bacteria that were present within crypts formed the interlaced layer before they mixed with the fecal compartment.
Interlaced layer The interlaced layer was mainly composed of the same bacterial groups that were present within the crypts (Table 3). These bacteria were condensed in extremely dense mats adjacent to the mucosa, which were clearly demarcated from the rest of the feces. This layer was relatively thin in WT mice and in IL-10 knockout mice without colitis. However, the interlaced layer was markedly amplified in all mice with colitis.
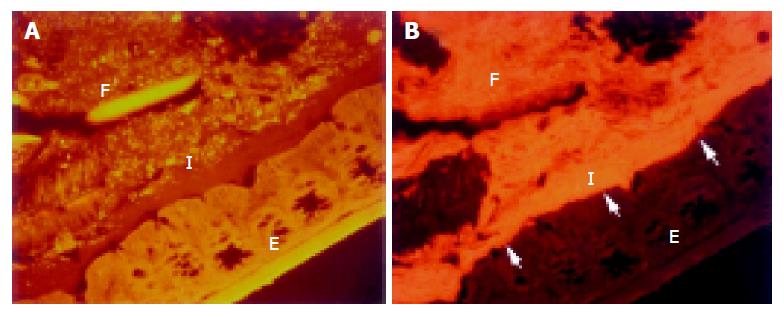
The concentration of bacteria into the interlaced layer was not simply numerical. The interlaced layer prevented the mucosa from contact with bacterial groups hybridizing with Bac303 (Bacteroides) and Clit135 (Clostridium difficile) probes which were observed in the cecum and initial parts of the proximal colon in healthy WT mice (Figure 6). The blocking role of the interlaced layer was especially well seen when pairs of probes representing all (Eub338)/interlaced (Erec, Alf1b, Phasco, Lab158, etc.) and exclusively fecal (e.g., Bac303, Clit135) bacterial population were simultaneously used (Figures 7, 8).
Adhesive bacteria Bacteria had direct contact with the cecal wall in healthy WT mice. These bacteria were separated completely from the wall when the fecal masses shrank during fixation without that bacteria adhered to the colonic surface (Figure 2B). Bacterial-mucosal contact in the cecum was therefore not adhesive.
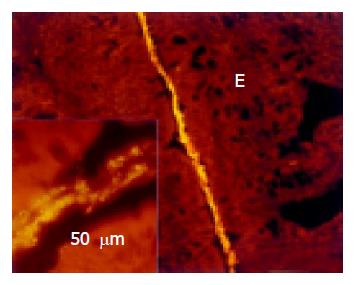
True adhesion was observed in all animals with colitis. This was characterized by bacteria, which lined the mucosal surface and were located beneath the mucus layer. This true adhesion was found in at least one location in all animals with colitis, but was observed mainly in the distal portion of the colon (Figure 9). Sixty percent of adhesive bacteria were Bacteroides.
Invading bacteria Bacteria invading the mucosa were found exclusively in the rectum and distal colon of DSS and IL-10 knockout mice with colitis (Figure 10). The types of invading bacteria were heterogeneous but mainly represented bacteria of the Bacteroides and Erec groups. The proportion of Bacteroides to Erec was more than three to one.
In the present study we described a novel approach for the characterization of biofilm in the murine intestine. The utilization of the broad in situ typing methodology, described herein, allowed for the first time to visualize the spatial complexity of murine intestinal microbiota in health and disease. Our results clearly show that normal murine intestinal flora is highly differentiated, organized in structurally definitive, distinctively composed compartments, which are typical for each colonic segment and closely interrelated to colonic function. Finally, we report, for the first time, that intestinal inflammation is associated with spatial re-distribution of specific groups of bacteria in the large intestine of mice.
In recent years a large amount of data has emerged that point out the importance of biofilm in environmental microbiology in such diverse settings as implantable medical devices[31] and dental plaque[32,33]. In regards to the intestinal microbiota, however, detailed investigations are clearly lacking. The limited number of studies that incorporated FISH technology to investigate microorganisms in histologic material of human or animal intestines has dealt for the most part with the detection and enumeration of bacteria[34-36]. In contrast, in the present study, we modified the existing methodology to achieve better characterization of the spatial association between intestinal bacteria and the mucosal environment. First, to accomplish optimal resolution of the spatial structures, our technique included probes, which have been shown to express low bacterial specificity taking into account some level of uncertainty in regard to the specificity of identification of bacteria and the sensitivity of enumeration studies. Second, we hybridized all probes that generated positive signals in multiple combinations to each other. Probes, which cross-hybridized with a subset of bacteria even when high stringency conditions were applied, were subsequently excluded from the evaluation.
A major finding of our study is the clear diversity observed between different intestinal segments in regards to the number of intraluminal bacteria and the localization of microorganisms in relation to intestinal wall. Specifically, in our study, bacteria were rare in the small intestine. On the contrary, with the exception of middle colon, all colonic segments were found to contain a large number of bacteria within the fecal mass. Striking differences, however, were observed between proximal and distal colonic areas regarding the position of microorganisms in relation to the bowel wall. Indeed, not only were the cecum and proximal colon tightly filled with high concentrations of bacteria, but direct contact of all involved bacterial groups with the bowel wall was observed as well. More importantly, abundant bacteria within colonic crypts were a uniform characteristic of proximal large intestinal segments. On the other hand, in the distal colon and rectum intraluminal bacteria were prohibited from contact to the bowel wall, whereas no microorganisms were detected in the colonic crypts.
We hypothesize that the differences in the spatial association between commensal bacteria and murine colonic wall replicate functional differences between individual colonic segments. First, our data indicate that the cecum and proximal colon, where direct contact of bacteria and bowel wall takes place, are involved in the proper fermentation reactor of the large bowel in mice. In addition, the rich bacterial population found within the crypts of proximal colonic segments of WT mice contradicts the previous assumption that such localization of bacteria indicates the presence of disturbed mucosal barrier[38]. A more plausible explanation is that these bacteria represent inocula used by the host to boost and maintain the stability of fermentation. This notion is strongly supported by the fact that intracryptic bacteria are devoid of potentially harmful species such as Bacteroides (Bac303) or Clostridium difficile (Clit135). Second, our findings support the concept that, different from the proximal colon, the distal colon functions mainly as a segregation device. In fact, the waste product segregation is managed by a mucus layer that normally starts in the middle colon and becomes fully developed in the distal colon, where the number of crypt bacteria continuously declines. The composition of mucins in the proximal and distal colonic segments in mice and men has been extensively studied and marked differences were previously described[37]. The lack of detection of a mucus layer by in situ hybridization in the cecum and proximal colon of WT mice does not indicate that mucin production by the cecal epithelium does not occur. It is more probable that mucus secreted in the murine cecum is penetrable by bacteria and does not lead to the development of the mucus layer, which physically separates the fecal masses from the epithelium. The “physiological” contact that takes place between bacteria and proximal large intestinal wall may underlie the tolerance of the colonic epithelium towards commensal flora that characterizes the healthy state. More importantly it offers an attractive explanation as to why colitis very often originates in the distal colon, where the bacteria are separated from the mucosa and tolerance does not develop. The functional differences between proximal and distal colon in healthy WT mice are further emphasized during peristalsis. The proximal and distal parts of the healthy WT mouse colon are dilated and filled with feces and divided by a narrow middle colon, which prevents the intermixing of the two reservoirs.
A unique finding in our study is the identification of the interlaced bacterial layer. This represents a bacterial sheet that is composed of selected microbial groups, which grow in dense mats between feces and mucosa and are impenetrable for most fecal bacteria. The interlaced layer is an entirely new entity observed in the transition zone between proximal and distal colon. The reason why previous studies failed to detect this distinctive layer may lie in the fact that the interlaced layer is relatively thin in healthy WT mice and poorly contrasts against the bright fluorescence of highly concentrated dense fecal compartment when universal probes or single group-specific probes are used. It can be definitively recognized only when pairs of probes specific for strictly fecal bacteria such as Bacteroides along with crypt-specific bacterial probes such as Erec are concomitantly applied, an approach that has not been practiced until now. The functional peculiarities of the interlaced layer indicate that it could be nature’s solution for preventing the spoiling of the fermentation by overgrowth of contaminants or facultative pathogens.
Our studies in DSS-treated and IL-10 knockout mice clearly demonstrate that, in the setting of intestinal inflammation, the described complex cooperative work of macro and microorganisms within the intestinal lumen was deranged. The development of colitis introduced changes that can be interpreted as increasing host intolerance to the fecal flora. First, in the inflamed colon the formation of the mucus layer that prevents contact of feces with the colonic wall moved proximally and became detectable in the cecum. Second, the spread and width of the mucus layer significantly increased throughout the colon. Third, redistribution of the intestinal microorganisms occurred that was highly selective and compartment-specific. Indeed, the concentrations of bacteria in the fecal compartment dropped dramatically. Nevertheless, while microorganisms were suppressed within the fecal compartment, the intra-cryptic bacterial groups recognized by the Alf1b, Erec, Phasco and Lab158 probes as well as the interlaced bacterial layer were enhanced in their growth. Both the number of crypts containing bacteria and the number of bacteria within single crypts significantly increased in mice with colitis. Furthermore, intra-cryptic bacteria were regularly found even in the distal colon. The interlaced layer got especially dense and thick in the inflamed proximal and middle colon and spread up into the cecum. Although these results indicate an activation of intestinal defense mechanisms in mice with colitis, as expressed by the wider spread and increased thickness of mucus and the interlaced layer, fecal bacteria are able to overcome the defense lines. In fact a definite discriminatory characteristic between healthy and colitic mice is the presence of invasive bacteria and bacteria that were adhesive to the mucosa below the mucus in the latter group. Specification analysis revealed that Eubacterium rectale group was most numerous in crypts, feces and the interlaced layer. On the other hand, Bacteroides species dominated the adhesive and invasive populations documenting the changing spectrum of bacteria involved in the inflammatory process.
In conclusion, the present study indicates that the proximal, middle and distal colonic segments appear to be physiologically distinct with respect to their interactions with bacteria. On the other hand, bacterial attachment to the mucosa can follow one of three principally different processes: fermentation (facilitated by direct contact to the proximal colonic wall), protection (through the formation of an isolating interlaced layer), and inflammation (adhesion and invasion). These processes are differently expressed throughout the colon and have opposite meanings in each case and position. The first is reduced, whereas the last two are enhanced in colitis. As an end result, the interrelation between bacteria and colonic epithelium and the state that predominates (tolerance vs. inflammation) is constantly defined by the mode of bacterial attachment and the colonic segment where this attachment takes place. The spatial, segment-specific structure and differential response of specific bacterial groups and compartments to inflammatory stimuli may be pivotal for the clarification of many past inconsistencies of experimental and clinical research regarding the pathogenic role of bacteria in IBD[38-41] and is a promising goal for future studies.
| 1. | Sartor RB. Pathogenesis and immune mechanisms of chronic inflammatory bowel diseases. Am J Gastroenterol. 1997;92:5S-11S. [PubMed] |
| 2. | Wilson M. Bacterial biofilms and human disease. Sci Prog. 2001;84:235-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 72] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 3. | Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2693] [Cited by in RCA: 2773] [Article Influence: 115.5] [Reference Citation Analysis (3)] |
| 4. | Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143-169. [PubMed] |
| 5. | Matsuo K, Ota H, Akamatsu T, Sugiyama A, Katsuyama T. Histochemistry of the surface mucous gel layer of the human colon. Gut. 1997;40:782-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 191] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 6. | Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762-770. [PubMed] |
| 7. | Manz W, Amann R, Ludwig W, Wagner M, Schleifer KH. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: Problems and solutions. System Appl Microbiol. 1992;15:593-600. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1470] [Cited by in RCA: 1220] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 8. | Bohnert J, Hübner B, Botzenhart K. Rapid identification of Enterobacteriaceae using a novel 23S rRNA-targeted oligonucleotide probe. Int J Hyg Environ Health. 2000;203:77-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Poulsen LK, Licht TR, Rang C, Krogfelt KA, Molin S. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J Bacteriol. 1995;177:5840-5845. [PubMed] |
| 10. | Trebesius K, Harmsen D, Rakin A, Schmelz J, Heesemann J. Development of rRNA-targeted PCR and in situ hybridization with fluorescently labelled oligonucleotides for detection of Yersinia species. J Clin Microbiol. 1998;36:2557-2564. [PubMed] |
| 11. | Amann RI, Stromley J, Devereux R, Key R, Stahl DA. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992;58:614-623. [PubMed] |
| 12. | Hristova KR, Mau M, Zheng D, Aminov RI, Mackie RI, Gaskins HR, Raskin L. Desulfotomaculum genus- and subgenus-specific 16S rRNA hybridization probes for environmental studies. Environ Microbiol. 2000;2:143-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 28] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 13. | Feydt-Schmidt A, Rüssmann H, Lehn N, Fischer A, Antoni I, Störk D, Koletzko S. Fluorescence in situ hybridization vs. epsilometer test for detection of clarithromycin-susceptible and clarithromycin-resistant Helicobacter pylori strains in gastric biopsies from children. Aliment Pharmacol Ther. 2002;16:2073-2079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 14. | Snaidr J, Amann R, Huber I, Ludwig W, Schleifer KH. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884-2896. [PubMed] |
| 15. | Roller C, Wagner M, Amann R, Ludwig W, Schleifer KH. In situ probing of gram-positive bacteria with high DNA G + C content using 23S rRNA-targeted oligonucleotides. Microbiology. 1994;140:2849-2858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 408] [Cited by in RCA: 339] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 16. | Meier H, Amann R, Ludwig W, Schleifer KH. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G + C content. Syst Appl Microbiol. 1999;22:186-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 215] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 17. | Urdaci MC, Regnault B, Grimont PA. Identification by in situ hybridization of segmented filamentous bacteria in the intestine of diarrheic rainbow trout (Oncorhynchus mykiss). Res Microbiol. 2001;152:67-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 18. | Franks AH, Harmsen HJ, Raangs GC, Jansen GJ, Schut F, Welling GW. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336-3345. [PubMed] |
| 19. | Harmsen HJ, Raangs GC, He T, Degener JE, Welling GW. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl Environ Microbiol. 2002;68:2982-2990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 272] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 20. | Harmsen HJ, Elfferich P, Schut F, Welling GW. A 16S rRNA-targeted probe for detection of lactobacilli and enterococci in fecal samples by fluorescent in situ hybridization. Microbiol Ecol Health Dis. 1999;11:3–12. [RCA] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 220] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 21. | Frahm E, Heiber I, Hoffmann S, Koob C, Meier H, Ludwig W, Amann R, Schleifer KH, Obst U. Application of 23S rDNA-targeted oligonucleotide probes specific for enterococci to water hygiene control. Syst Appl Microbiol. 1998;21:450-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 33] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 22. | Jansen GJ, Mooibroek M, Idema J, Harmsen HJ, Welling GW, Degener JE. Rapid identification of bacteria in blood cultures by using fluorescently labeled oligonucleotide probes. J Clin Microbiol. 2000;38:814-817. [PubMed] |
| 23. | Harmsen HJ, Wildeboer-Veloo AC, Grijpstra J, Knol J, Degener JE, Welling GW. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl Environ Microbiol. 2000;66:4523-4527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 238] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 24. | Zoetendal EG, Ben-Amor K, Harmsen HJ, Schut F, Akkermans AD, de Vos WM. Quantification of uncultured Ruminococcus obeum-like bacteria in human fecal samples by fluorescent in situ hybridization and flow cytometry using 16S rRNA-targeted probes. Appl Environ Microbiol. 2002;68:4225-4232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 91] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 25. | Jensen TK, Boye M, Ahrens P, Korsager B, Teglbjaerg PS, Lindboe CF, Møller K. Diagnostic examination of human intestinal spirochetosis by fluorescent in situ hybridization for Brachyspira aalborgi, Brachyspira pilosicoli, and other species of the genus Brachyspira (Serpulina). J Clin Microbiol. 2001;39:4111-4118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 26. | Langendijk PS, Schut F, Jansen GJ, Raangs GC, Kamphuis GR, Wilkinson MH, Welling GW. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;61:3069-3075. [PubMed] |
| 27. | Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer KH. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology. 1996;142:1097-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 889] [Cited by in RCA: 786] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 28. | Suau A, Rochet V, Sghir A, Gramet G, Brewaeys S, Sutren M, Rigottier-Gois L, Doré J. Fusobacterium prausnitzii and related species represent a dominant group within the human fecal flora. Syst Appl Microbiol. 2001;24:139-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 133] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 29. | Manz W, Eisenbrecher M, Neu TR, Szewzyk U. Abundance and spatial organization of gram-negative sulfatereducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol Ecol. 1998;25:43-61. [RCA] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 150] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 30. | Stahl DA, Amann R. Development and application of nucleic acid probes in bacterial systematics. In E. Stackebrandt and M.Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley Sons Ltd. Pub 1991; 205–248. |
| 31. | Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest. 2003;112:1466-1477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 504] [Cited by in RCA: 446] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 32. | Kolenbrander PE. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol. 2000;54:413-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 593] [Cited by in RCA: 580] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 33. | Marsh PD, Bradshaw DJ. Dental plaque as a biofilm. J Ind Microbiol. 1995;15:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 234] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 34. | Schultsz C, Van Den Berg FM, Ten Kate FW, Tytgat GN, Dankert J. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology. 1999;117:1089-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 207] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 35. | Kleessen B, Kroesen AJ, Buhr HJ, Blaut M. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand J Gastroenterol. 2002;37:1034-1041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 249] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 36. | Kleessen B, Hartmann L, Blaut M. Fructans in the diet cause alterations of intestinal mucosal architecture, released mucins and mucosa-associated bifidobacteria in gnotobiotic rats. Br J Nutr. 2003;89:597-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 135] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 37. | Einerhand AW, Renes IB, Makkink MK, van der Sluis M, Büller HA, Dekker J. Role of mucins in inflammatory bowel disease: important lessons from experimental models. Eur J Gastroenterol Hepatol. 2002;14:757-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 104] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 38. | Campieri M, Gionchetti P. Bacteria as the cause of ulcerative colitis. Gut. 2001;48:132-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 144] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 39. | Sartor RB. Microbial factors in the pathogenesis of Crohn's disease, ulcerative colitis, and experimental intestinal inflammation. Philadelphia: Saunders Pub 2000; 153-178. |
| 40. | Guarner F, Malagelada JR. Role of bacteria in experimental colitis. Best Pract Res Clin Gastroenterol. 2003;17:793-804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 66] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 41. | Shanahan F. Inflammatory bowel disease: immunodiagnostics, immunotherapeutics, and ecotherapeutics. Gastroenterology. 2001;120:622-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 243] [Article Influence: 9.7] [Reference Citation Analysis (0)] |