Published online Dec 28, 2005. doi: 10.3748/wjg.v11.i48.7579
Revised: July 10, 2005
Accepted: July 15, 2005
Published online: December 28, 2005
AIM: To develop a simplified and efficient method for the preparation of hepatitis C virus (HCV) cDNA microarray probes.
METHODS: With the technique of restriction display PCR (RD-PCR), restriction enzyme Sau3A I was chosen to digest the full-length HCV cDNAs. The products were classified and re-amplified by RD-PCR. We separated the differential genes by polyacrylamide gel electrophoresis and silver staining. Single bands cut out from the polyacrylamide gel were isolated. The third-round PCR was performed using the single bands as PCR template. The RD-PCR fragments were purified and cloned into the pMD18-T vector. The recombinant plasmids were extracted from positive clones, and the target gene fragments were sequenced. The cDNA microarray was prepared by spotting RD-PCR products to the surface of amino-modified glass slides using a robot. We validated the detection of microarray by hybridization and sequence analysis.
RESULTS: A total of 24 different cDNA fragments ranging from 200 to 800 bp were isolated and sequenced, which were the specific gene fragments of HCV. These fragments could be further used as probes in microarray preparation. The diagnostic capability of the microarray was evaluated after the washing and scanning steps. The results of hybridization and sequence analysis showed that the specificity, sensitivity, accuracy, reproducibility, and linearity in detecting HCV RNA were satisfactory.
CONCLUSION: The RD-PCR technique is of great value in obtaining a large number of size-comparable gene probes, which provides a speedy protocol in generating probes for the preparation of microarrays. Microarray prepared as such could be further optimized and applied in the clinical diagnosis of HCV.
- Citation: Sun ZH, Ma WL, Zhang B, Peng YF, Zheng WL. Application of restriction display PCR technique in the preparation of cDNA microarray probes. World J Gastroenterol 2005; 11(48): 7579-7584
- URL: https://www.wjgnet.com/1007-9327/full/v11/i48/7579.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i48.7579
Hepatitis C virus (HCV) is a RNA virus with a high rate of genetic mutation[1]. HCV has a linear genome approximately 10 kb in length, which consists of a positive sense single-stranded RNA (ssRNA)[2]. Consistent with related members of the family Flaviviridae, HCV demonstrates a high degree of sequence variation throughout its genome. Sequence analysis of multiple strains of HCV has demonstrated that the nucleotide sequence can differ by as high as 30%[3]. Infection with HCV has been identified as the major cause of post-transfusion non-A, non-B hepatitis, and is a major public health problem in most areas of the world, raising the issues of its diagnosis, treatment, and prevention[4-6].
In the present study, we utilized the technique of restriction display PCR (RD-PCR) to prepare HCV cDNA chip probes. Restriction enzyme Sau3A I was chosen to digest the full-length HCV cDNAs. The products were classified and re-amplified by RD-PCR. We separated the differential genes by polyacrylamide gel electrophoresis and silver staining. The cDNA microarray was prepared by spotting RD-PCR products to the surface of amino-modified glass slides using a robot. The specificity, sensitivity, accuracy, reproducibility, and linearity in detecting HCV RNA were evaluated.
The full-length plasmid of HCV pCV-J4L6S was presented by Dr Jens Bukh of NIH (USA)[7].
Premix Taq, dNTP, EcoRI, NotI, XbaI, Sau3A I, pMD18-T vector, and T4 DNA ligase were obtained from Takara Corp. (Japan). Dimethyl sulfoxide (DMSO) was bought from Sangon (Shanghai, China). Plasmid Miniprep kits, 3S PCR purification kit V20 were purchased from Shen Neng Bo Cai Corp. (China). PCR primers of HCV and the primers in pMD18-T vector were synthesized by BioAsia Corp. (China). Universal primer (U) cy3-GTTTG GCTGGTGT GGATC, selective primers (similar with U but with one “nesting” base overhanging at 3’-end, UA, UT, UC, UG) and the adapter (SIP, SIR) were purchased from GIBCO Corp. (USA).
GenePix 4000B scanner and ScanArray Lite were provided by GSI Lumonics (Billerica, USA). GS Gene Linker ultraviolet chamber was obtained from BioRad (Hercules, USA). PixSys 5500 gene chip printing machine was purchased from Cartesian Technologies (Irvine, USA). CMT-GAPSTM coated slides and Corning CMT-HybridizationTM chambers were ordered from Corning Microarray Technology (Acton, USA).
The E. coli strain XL-1 used in experiments was maintained in our laboratory.
To isolate HCV genomes, plasmid pCV-J4L6S was digested with NotI and XbaI. The plasmid (1-2 μg) was added to a total volume of 20 μL mixture at 37 °C for 4 h. The target HCV gene was isolated and recovered with UNIQ-5 column DNA extraction kit (Sangon, China). RD-PCR was performed as previously described[8-10]. The recovered HCV cDNAs were digested with 2 μL Sau3A1 (5′↓GATC3′) (10 U/μL) in a total volume of 20 μL mixture at 37 °C for 3 h. The two ends of each Sau3AI-digested fragment were linked to an adapter that was prepared by annealing the 2 oligonucleotides containing the sequences of 5’-GATCCACACCAGCCAAACC CA (SIP) and 5’-GGTTTGGCTGGTGTG (SIR) in a ligation reaction containing 1 μL T4 DNA ligase (350 U/μL), 1 μL 10×DNA ligation buffer, 1 μL adapter (50 μmol/L), 20 pmol of digested HCV cDNA fragments of Sau3AI, and then UPW was added for a total of 10 μL. After 4 h of ligation at 16 °C, PCR was performed in a 9700 thermocycler with an initial denaturation at 94 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, at 60 °C for 30 s, at 72 °C for 1 min, and a final extension at 72 °C for 7 min. PCR primers were designed to match the universal adapters, including the restriction site sequence, but with one “nesting” base overhanging at the 3’-end, and the reactions were divided into 10 subgroups. To check for positive PCR results, 7 μL of the PCR products was loaded onto 5% polyacrylamide (Takara, Japan) gel electrophoresis at 90 V for 5-6 h and the technique of DNA silver staining[11] was used to separate different target gene fragments.
The final concentration of each probe was adjusted to 0.3 mg/mL with DMSO and water. The DMSO concentration was 50% (v/v). The probes were spotted onto a CMT-GAPS aminosilane-coated glass microscope slide at 25 °C and 60% relative humidity, using the ArrayIt ChipMaker2TM microspotting pins (TeleChem International, Sunnyvale, USA) and a Cartesian PixSys 5500 robot. A microarray of 12×15 spots was printed. The arrangement of all spots on the gene chip is shown in Figure 1. After printing, the slide was rehybridized and snap-dried in a plate at 80 °C for 5 min. A BioRad UV crosslinker was used to immobilize the DNAs onto the slide with 125 mJ of energy. The slide was treated with blocking solution (335 mL 1-methyl-2-pyrrolidinone, 6 g succinic anhydride and 15 mL 1 mol/L sodium borate, pH 8) and stored for later use.
The samples of HCV were digested at 37 °C with the restriction enzyme Sau3AI. T4 DNA ligase was used to link gene fragments with universal adapters (SIP, SIR). After 3 h of ligation at 16 °C, 25 μL 2×premix, 2 μL of linkage products, 2 μL of Cy3 labeled universal primer matched with the adapter, and 21 μL of water were added for a total volume of 50 μL. Reactions were performed in a GeneAmp PCR 9700 system with an initial denaturation at 94 °C for 10 min, and then subjected to 30 thermal cycles at 94 °C for 30 s, at 60 °C for 30 s, at 72 °C for 1 min, followed by a final incubation at 72 °C for 5 min. Finally, the reaction was stopped by cooling down the solution to 4 °C. After this labeling reaction, the sample DNA fragments of hundreds of base pairs were labeled with fluorescent Cy3. The labeled PCR products were further purified using a 3S PCR product purification kit V20. Of the 30 μL purified products, 5 μL was used for hybridization with the microarray probes.
The slide was incubated in 25% formamide, 5×SSC, 0.1% sodium dodecyl sulfate (SDS) in a jar for 45 min at 42 °C, rinsed 5 times in distilled water, immersed in isopropyl alcohol for 1 s, and then dried in the air. Five microliters of Cy3 labeled samples was mixed with 1 μL Cot-1 DNA (20 μg/μL, Life Technologies) and 6 μL 2×hybridization buffer (50% formamide, 10×SSC, 0.2% SDS) that was preheated at 42 °C, then heated to 95 °C for 5 min and centrifuged at 14 000 g for 2 min. This mixture was completely pipetted onto the pre-hybridized lambda phage DNA microarray slide and covered with a glass coverslip pretreated with Sigmacote® (Sigma, St. Louis, USA) to keep the sample from evaporating. The slide was put into a sealed hybridization box, 10 μL UPW was added to each well at the two ends of the chamber. The sealed chamber containing the DNA microarray slide was placed in a 42 °C water bath. After 4 h of incubation, the slide was taken out and washed in low-stringency washing buffer containing 1×SSC and 0.2% SDS at 42 °C for 5 min, in high-stringency washing buffer containing 0.1×SSC and 0.2% SDS at room temperature for 10 min, in 0.1×SSC and Milli Q water and ethanol, respectively. Finally, the air-dried slide was scanned using the ScanArray® Lite MicroArray Analysis System.
The hybridized microarrays were scanned using GenePix 4000B scanner under the conditions of 90% laser power and 70% photo-multiplier tube (PMT). The results were analyzed using the QuantArray array analysis software. The criteria for positivity included the average fluorescence signal of Cy3 being three times as great as the value of the negative point, with a retro value of 2.7-3.3.
Ten HCV gene fragments were selected from the high value of the hybridized fluorescence signals to low fluorescence signals as microarray probes. The probes were spotted onto a CMT-GAPS aminosilane-coated glass microscope slide at 25 °C and 60% relative humidity, using the ArrayIt ChipMaker2TM microspotting pins and a Cartesian PixSys 5500 robot. A microarray of 12×8 spots was printed. The arrangement of all spots on the gene chip was similar to that shown in Figure 1. After printing, the slide was rehybridized and snap-dried in a plate at 80 °C for 5 min. A BioRad UV crosslinker was used to immobilize the DNAs onto the slide with 125 mJ of energy. The slide was treated with blocking solution and stored for later use.
The specificity, sensitivity, accuracy, reproducibility, and linearity of this assay system were evaluated.
After purification using the 3S PCR purification kit V20, the RD-PCR products (for probe preparation) and real-time RT-PCR products (for clinical serum sample detection) were inserted into the pMD18-T vector. The ligation mixture containing 4 μL of PCR products, 1 μL of pMD18-T vector (50 ng/μL) and 5 μL of loading buffer solution was incubated at 16 °C for 3 h, and then transferred into 100 μL of XL-1 E. coli competent cells treated with solutions containing Ca2+ ions (0.1 mol/L). To transform E. coli, the mixture of DNA formed in a ligation reaction was combined with a suspension of competent cells for 30 min, then heat-shocked at 42 °C for 1-2 min. The cells were then incubated in a growth medium and finally spread on an agar plate and incubated until single bacterial colonies were grown. Then the clones containing target fragments were selected and identified with the pMD18-T vector primer (primer A 5′-GTAAAACGACGGCC AGT-3′, primer B 5′-CAGGAAACAGCTATGAC-3′). To check for positive PCR results, 5 μL of the PCR products was analyzed by 1.5% agarose (Takara, Japan) gel electrophoresis at 75 V for 45 min with a DNA marker DL2000 (Takara, Japan) as reference. Then the sequence was analyzed with ABI PrismTM 3730 DNA sequencer, and GenBank Blast sequence alignments were done.
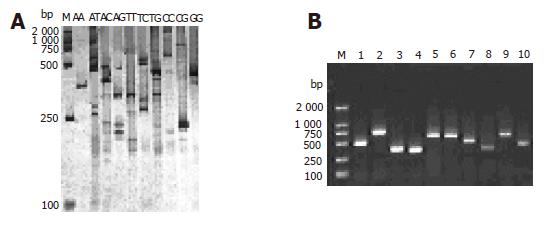
Twenty-four gene fragments of HCV were found in each subgroup by RD-PCR amplification with the expected size. The products were separated by electrophoresis on 5% polyacrylamide gel, and cDNA bands were stained with a silver solution (Figure 2A). Each subgroup produced 1-5 single cDNA bands with their length ranging from 200 to 800 bp. These bands could be used as probes for chip manufacture. Figure 2B shows the 1.5% agarose gel electrophoresis results of the clones.
The clinical diagnostic microarray was prepared by immobilizing the captured target genes of pathogens on a slide specifically treated. The DNA or RNA extracted from the patient’s serum was labeled with fluorochrome and hybridized to the target DNA. In this study, the microarray was prepared by spotting RD-PCR products of HCV onto the surfaces of glass slides using a Cartesian 5500 MicroArrayer. Controls were immobilized at the same time. The control system was composed of empty controls which were DMSO without gene fragments, negative controls which were gene fragments of plants (rice) (B1-6) and an eukaryocyte (K562 cell) (B7-12) and a prokaryocyte (E. coli) (C1-6) not homologous with HCV, positive controls which were the reconstructed gene fragments. Eight of the 12×15 microarrays could be simultaneously immobilized on a glass slide.
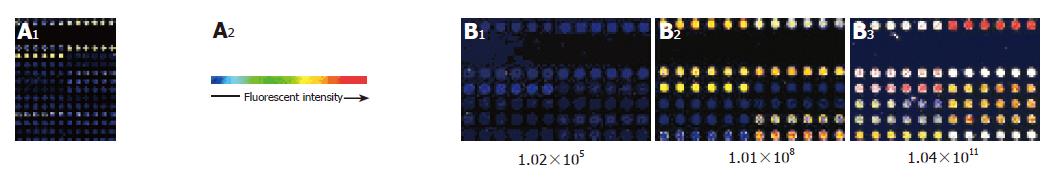
Based on the hybridizing signals under 90% laser energy and 70% GMT on the gene chip, the specificity and sensitivity in detecting HCV were satisfactory (Figure 3A). There was no signal on the empty control spots (printing 50% DMSO) and negative control spots (printing gene fragments of eukaryotic cells). The signal of spots hybridized to HCV positive controls and samples of HCV was strong and clear, but the analysis of variance showed that the density of signal was different in different probes (F = 8.325, P<0.001).
Ten HCV gene fragments selected from the high- to low-density signals of hybridization were prepared for HCV gene chip probes. To obtain further information about the ten probes, we sequenced and analyzed the probes and found that nearly all of them had a similar length ranging 250-700 bp and a high GC content (≥40%) and Tm values (≥82 °C). The microarray was prepared by spotting the probes with Cartesian 5500 MicroArrayer. A microarray of 12×8 spots was printed. The arrangement of all spots on the gene chip is shown in Figure 1. The hybridization results indicated that the positive signal was strong compared to the hybridization signal (Figure 3B). There was no signal on the empty control spots and negative control spots.
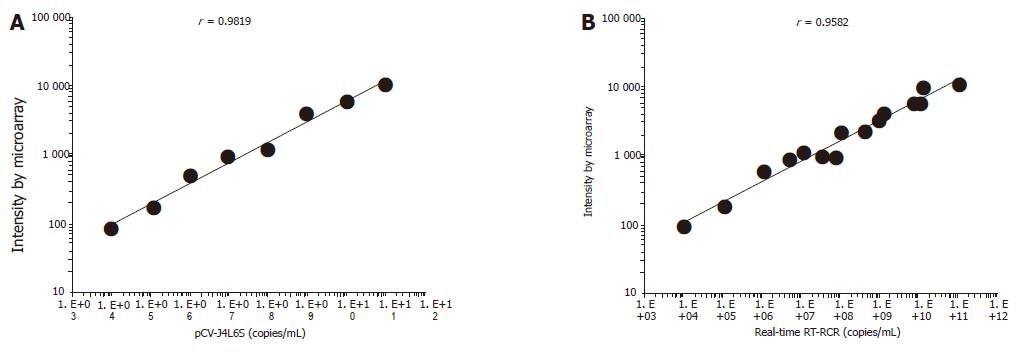
There was a strong linear relation between the concentrations of target cDNA and the fluorescence intensities obtained from microarray assay (r = 0.9819). The detection range of the microarray was 104-1011 copies/mL. The lower detection limit of HCV cDNA by microarray was 1.03×104 copies/mL, which was 2 log units lower than that by real-time RT-PCR. The reproducibility and accuracy of this assay system were evaluated by repeated measurement, and the within-run coefficient of validation was 6.6%, while the between-run coefficient of validation was 7.6%. Seventeen serum samples from hepatitis C patients (positive for anti-HCV, ALT≥80) were analyzed, and 15 patients (88.2%) were positive by microarray assay. Ten serum samples from healthy people were also evaluated and the results were all negative. The obtained sequences were verified and each sequenced PCR product was confirmed to be a HCV genome fragment using the basic local alignment search tool (BLAST) and the GenBank database.
HCV is the most important cause of transfusion-associated and community-acquired non-A, non-B hepatitis. Chronically infected individuals have a relatively high risk of developing chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. No effective vaccine therapy is available at present for HCV. Detection of HCV is essential for the correct diagnosis of HCV infection. Antibody-based methods have been considered as a practical way to detect infection of HCV. However, these methods cannot diagnose patients with hypoimmunity. Nucleic acid hybridization may have an excellent specificity, but its sensitivity is not satisfactory. In the protocol of PCR, cross contamination and false negative and positive incidents often occur. DNA microarray offers a solution to these problems and has the potential for the diagnosis of HCV infection. Compared to traditional diagnostic techniques, it has a number of advantages such as integration, micromation, and automatization[12,13]. The use of multiple independent gene fragments with a suitable size (ranging from 200 to 800 bp) for the probes to detect the same molecular targets can greatly enhance the signal-to-noise ratio and reduce the false-positive rate[14]. The gene chip also offers a dependable basis for the diagnosis and treatment of hepatotropic viral infections[15-17].
Probe preparation is a key step for microarray. One of the major difficulties in the development of microarray is to collect or prepare sufficient probes[18]. A few methods can be used to prepare microarray probes, including PCR amplification of DNA fragments with a molecular clone[19], artificial synthesis of oligonucleotide arrays by a DNA synthesizing machine[20] and light direction of in situ synthesis[19]. We prefer the first method because of its rapidity, simplicity, and effectiveness. However, conventional PCR is conducted with specific primers. RD-PCR provides an efficient and a simple way to obtain probes, since cDNA gene fragments digested by 4-cutter restriction endonuclease can be ligated with adapters at the same restriction site. In the present study, a PCR universal primer (U) was designed to match the sequence of both the adapters and the restriction site, but there were too many fragments amplified with universal primers (U). Using the selective primers with one or more “nesting” bases at the 3′-end of universal primer, 10 or more subgroups of PCR reaction were performed for various single selective primers or primer combination, in which gene fragments were widely distributed in different groups and their isolation was achieved. We separated the differential genes by polyacrylamide gel electrophoresis and silver staining. Single bands cut out from polyacrylamide gel were isolated. Using the single bands as PCR template, we performed the third-round PCR and a total of 24 different cDNA fragments ranging from 200 to 800 bp were obtained and sequenced, which were the specific gene fragments of HCV. These fragments could be further used as probes in microarray preparation, suggesting that RD-PCR technique is of great value in obtaining a large number of size-comparable gene probes, thus providing a swift protocol in generating probes for the preparation of microarrays.
In general, the density of hybridization signal is correlative to the length of probes, GC contents and Tm value under the same experimental conditions. A further sequence analysis of the 10 probes, GC contents, and Tm value showed that these probes were 250-700 bp in length, and had a high GC content (≥40%) and Tm values (≥82 °C).The probes were widely distributed in the full HCV genome. The results of hybridization and sequence analysis showed that the specificity, sensitivity, accuracy, reproducibility, and linearity in detecting HCV RNA were satisfactory. There was a linear correlation between the concentrations of pCV-J4L6S target cDNA and the signal intensities (correlation coefficient = 0.9819). The detection range of the microarray was 104-1011 copies/mL. The lower detection limit of HCV RNA was 1.03×104 copies/mL, which was 2 log higher than that by RT-PCR (Taqman method). But the assay was as sensitive as the conventional RT-PCR in HCV detection. The chip we developed is cost-effective, and the procedure required to prepare the chip is straightforward and convenient. When results are verified by molecular hybridization, PCR cross contamination can be overcome. By incorporating negative and positive controls, the detection results can be ensured. Subjective factors in terms of judging the results can be reduced greatly using analytic software[20]. Seventeen serum samples from hepatitis C patients (positive for anti-HCV, ALT≥80) were analyzed, and fifteen patients (88.2%) were positive by microarray assay. Within the detection range of 104-1011 copies/mL, there was a good correlation between the two assay systems (n = 15, r = 0.9582) (Figure 4B). Ten serum samples from healthy people were also detected by this assay and no specific signal intensities were obtained from the samples. Finally, the reproducibility and accuracy of this assay system were evaluated by repeated measurement, and the within-run coefficient of validation was 6.6%, while the between-run coefficient of validation was 7.6%. However, further work should be done. It took about 10 h to complete our assay. Modifying the DNA extraction procedure could reduce this time. The sensitivity of the assay could be enhanced by increasing the amount of captured cDNA on the slide, or by pretreatment of the sample RNA. A large number of serum samples should be tested to verify the reliability of microarray assay in the detection of HCV.
In conclusion, cDNA microarray technology can be applied to other pathogens and is a useful diagnostic method for HCV infection.
The authors thank Dr Jens Bukh for the presentation of HCV plasmid and Drs Zhang Bao and Shi Rong for critical technical assistance.
| 1. | Ogata N, Alter HJ, Miller RH, Purcell RH. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:3392-3396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 400] [Cited by in RCA: 401] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 2. | Farci P. Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome [Science 1989; 244: 359-362]. J Hepatol. 2002;36:582-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4996] [Cited by in RCA: 4670] [Article Influence: 126.2] [Reference Citation Analysis (1)] |
| 3. | Simmonds P. Variability of hepatitis C virus. Hepatology. 1995;21:570-583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 350] [Cited by in RCA: 330] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 4. | Alter HJ, Purcell RH, Shih JW, Melpolder JC, Houghton M, Choo QL, Kuo G. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med. 1989;321:1494-1500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1225] [Cited by in RCA: 1146] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 5. | Hoofnagle JH, di Bisceglie AM. The treatment of chronic viral hepatitis. N Engl J Med. 1997;336:347-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 715] [Cited by in RCA: 681] [Article Influence: 23.5] [Reference Citation Analysis (3)] |
| 6. | Cui J, Dong BW, Liang P, Yu XL, Yu DJ. Construction and clinical significance of a predictive system for prognosis of hepatocellular carcinoma. World J Gastroenterol. 2005;11:3027-3033. [PubMed] |
| 7. | Yanagi M, Purcell RH, Emerson SU, Bukh J. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology. 1999;262:250-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 153] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 8. | Ma WL, Zheng WL. The research and development of DNA microarray technology. Sci Found China. 1999;13:270-273. |
| 9. | Ma WL, Zheng WL, James FB. RD-PCR: A new technique of differential display.In: Sun yixian(ed).The development of biochemistry and molecular biology in PLA.vol.1. Beijing: Uniform Medical Publishing. 1998;99-113. |
| 10. | Zheng WL, Ma WL, Waes CV. The differential display of poly A polymerase in tumor cells of differential malignancy. In: Ye XS(ed). Investigation on cell modulation. vol.1. Beijing: Uniform Medical Publishinghouse. 1998;73-79. |
| 11. | Zhao F, Zhang SJ, Jiang SH. Modification of the technique of staining and the PAGE. J Clin Exp Pathol. 1999;15:401-402. |
| 12. | Sun ZH, Ma WL, Zheng WL. Microarrays development in the diagnosis of HBV and HCV. Med J Chin PLA. 2003;28:375-376. |
| 13. | Sun ZH, Zheng WL, Ma WL. The development of molecular diagnosis of viral hepatitis. Guangdong med J. 2003;24:440-442. |
| 14. | Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21:20-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1512] [Cited by in RCA: 1267] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 15. | Petrik J. Microarray technology: the future of blood testing? Vox Sang. 2001;80:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 53] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 16. | Zhaohui S, Wenling Z, Bao Z, Rong S, Wenli M. Microarrays for the detection of HBV and HDV. J Biochem Mol Biol. 2004;37:546-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 17. | Livache T, Fouque B, Roget A, Marchand J, Bidan G, Téoule R, Mathis G. Polypyrrole DNA chip on a silicon device: example of hepatitis C virus genotyping. Anal Biochem. 1998;255:188-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 97] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Stewart DJ. Making and using DNA microarrays: a short course at Cold Spring Harbor Laboratory. Genome Res. 2000;10:1-3. [PubMed] |
| 19. | Sun ZH, Zheng WL, Mao XM, Zhang B, Lü L, Ma XD, Shi R, Ma WL. Rapid preparation of DNA microarray using PCR for hepatitis B and D virus detection. Di Yi Jun Yi Da Xue Xue Bao. 2003;23:677-679. [PubMed] |
| 20. | Wang HY, Malek RL, Kwitek AE, Greene AS, Luu TV, Behbahani B, Frank B, Quackenbush J, Lee NH. Assessing unmodified 70-mer oligonucleotide probe performance on glass-slide microarrays. Genome Biol. 2003;4:R5. [PubMed] |
Co-first-authors: Wen-Li Ma and Zhao-Hui Sun
Science Editors Wang XL and Guo SY Language Editor Elsevier HK