Published online Dec 28, 2005. doi: 10.3748/wjg.v11.i48.7564
Revised: January 1, 2005
Accepted: January 5, 2005
Published online: December 28, 2005
AIM: To compare the gene expression profile in a pair of HBV-infected twins.
METHODS: The gene expression profile was compared in a pair of HBV-infected twins.
RESULTS: The twins displayed different disease outcomes. One acquired natural immunity against HBV, whereas the other became a chronic HBV carrier. Eighty-eight and forty-six genes were found to be up- or down-regulated in their PBMCs, respectively. Tumor necrosis factor-alpha-induced protein 1 (TNF-αIP1) that expressed at a higher level in the HBV-immune twins was identified and four pairs of siblings with HBV immunity by RT-PCR. However, upon HBV core antigen stimulation, TNF-αIP1 was downregulated in PBMCs from subjects with immunity, whereas it was slightly upregulated in HBV carriers. Bioinformatics analysis revealed a K+ channel tetramerization domain in TNF-αIP1 that shares a significant homology with some human, mouse, and C elegan proteins.
CONCLUSION: TNF-αIP1 may play a role in the innate immunity against HBV.
- Citation: Lin MC, Lee NP, Zheng N, Yang PH, Wong OG, Kung HF, Hui CK, Luk JM, Lau GKK. Tumor necrosis factor-α-induced protein 1 and immunity to hepatitis B virus. World J Gastroenterol 2005; 11(48): 7564-7568
- URL: https://www.wjgnet.com/1007-9327/full/v11/i48/7564.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i48.7564
Though substantial advances have been made in understanding of the pathogenesis of hepatitis B virus (HBV), HBV infection remains a global health threat with currently more than 350 million carriers and causes about one million deaths worldwide. The situation is particularly severe in high-endemic areas like southeastern Asia. For instance, a study of 16 334 subjects in 1978-1979 in Hong Kong suggested that 43% of the local population have evidence of past infection and 10% are HBs Ag carriers[1].
The major spread mode of HBV in most high endemic areas, such as Hong Kong and China, is perinatal transmission, which accounts for 40%-50% of chronic HBV infection[2,3]. The reason for the preponderance of perinatal transmission among Orientals is at least in part related to the high prevalence of HBV infection among the Asian carriers of reproductive age[2]. Though the incidence of perinatal transmission is high in China, not all siblings in the same family with HBV-infected mothers remain persistently infected with HBV as some acquired natural immunity against the virus. We are among the first group to demonstrate that the use of HLA-matched donor marrow from siblings with natural immunity could enable serological clearance of HBV in their HBsAg positive recipient siblings[4-7]. This indicates that the transfer of certain molecules from the donor’s immune system is sufficient to confer the recipient immunity against HBV. However, the identities of these molecules are unknown.
In this study, Affymetrix cDNA mircroarray was employed to investigate the differential gene expression patterns in PBMCs in an identical twin pair who had different outcomes to HBV infection. Use of the twin pair could eliminate the potential confounding factors like age, gender, and living environment, etc. A novel cytokine signaling-related gene, TNF-αIP1, is significantly downregulated in chronic HBV carriers. Moreover, we have confirmed that this gene is indeed differentially expressed in several groups of HBV-infected siblings who display different disease outcomes by reverse transcription-polymerase chain reaction (RT-PCR). We also found that upon HBcAg stimulation, TNF-αIP1 exhibited different responses between PBMC isolated from subjects with immunity and chronic infection. These results suggest that TNF-αIP1 may be involved in immunity against HBV infection. An understanding of the immunological and genetic differences in PBMCs of these sibling pairs should provide insight into the molecular mechanisms of the protection and enable a more rationale design of therapeutic regimen for chronic HBV infection in Chinese.
Eight groups of HLA-A, B and DR identical Chinese siblings (including pair of identical twins) who differed in their outcomes to HBV infection were used in this study. RNA was isolated from PBMC of the siblings with spontaneous recovery (anti-HBs and anti-HBc positive) and their corresponding HLA-matched HBV-infected siblings (HBsAg positive).
Heparinized venous blood was collected and PBMC was separated by density-gradient centrifugation over Ficoll-Hypaque. Total RNA was extracted from PBMC using RNeasy kit (Qiagen, Valencia, CA, USA) following manufacturer’s instructions. For each sample, 60 mg of total RNA was reverse transcribed using Superscript II (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instructions.
Generation of antisense RNA (aRNA) probe for cDNA microarray analysis was carried out as described previously[8]. Briefly, mRNA was amplified using two or three rounds of cDNA synthesis followed by aRNA synthesis. T7-promoter-oligo-dT [5’-GCCAGTGAATTGTAATAC GACTCACTATAGGGAGGCGG-(dT)24-3’] was used for cDNA synthesis and T7 RNA polymerase was used for aRNA synthesis. Ten micrograms of starting total RNA was used to obtain 10 mg of final aRNA. During the process of the final aRNA synthesis, biotinylated UTP and CTP were used for labeling purpose (Enzo, Farmingdale, NY, USA). Detection of the hybridized probe using streptavidin-phycoerythrin fluorescent conjugate was done according to manufacturer’s protocol (Molecular Probes, Eugene, OR, USA).
The labeled aRNA probe from PBMC was hybridized to a HU 95A gene chip representing 12 000 full-length human genes (Affymetrix, Santa Clara, CA, USA). The threshold for significant up- or down-regulation was 2.0- and 0.5-fold, respectively. Hybridizations were typically carried out for 16 h at 45 °C followed by washing, staining and using Affymetrix fluidic stations. Stained arrays were scanned in the G2500 A Hewlett-Packard Gene Array Scanner (Hewlett-Packard, Palo Alto, CA, USA) at the excitation wavelength of 488 nm. The amount of emitted light was proportional to the bound target at each location on the probe arrays.
Total RNA was reverse transcribed using the Superscript II (Invitrogen) according to the manufacturer’s instructions. PCR conditions for pre-B-cell colony-enhancing factor (PBEF), IL-18 receptor accessory protein (IL-18 RAP), TNF-αIP1 and GADPH were as follows: at 94 °C for 30 s, at 57 °C for 30 s, at 72 °C for 1 min for 35, 35, 35, and 32 cycles, respectively. The primer sequences of the genes tested were PBEF (sense: 5’-AAAAGCTGTTCCTGAGGGCTTTG-3’; anti-sense: 5’-TGACCACAGATACAGGCACTGATG-3’); IL-18RAP (sense 5’-CCAGAGCCACAGAAATCACATTTC-3’; anti-sense 5’-CAAGAAATAGAGCCAGTGCTCCCA-3’); TNF-αIP1 (sense: 5’- TTACCTCCGAGATGACACCATCAC-3’; anti-sense: 5’-TCCTCATCTTCACTGGGGGAA-3’).
Perinatal transmission is the most common spread mode of hepatitis B infection in Chinese. Yet not all the siblings infected with HBV from mothers become chronic HBV carriers. It appears that different disease outcomes of these siblings represent success and failure of their immune systems in controlling HBV infection. In this study, we attempted to explore this phenomenon by investigating gene expression profiles in PBMCs of two identical twins who displayed diverse disease outcomes.
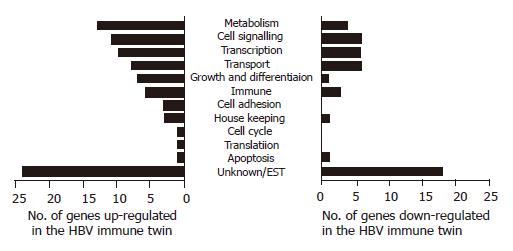
The differences in PBMC gene expression between two identical twins were analyzed using a DNA microarray chip containing 12 000 human expressed genes. Eighty-eight genes were expressed at a higher level and 46 genes were expressed at a lower level in twins with immunity (HBV resistant) compared to the HBV carrier twins [HBV susceptible). These genes were grouped on the basis of their predicted functions into 12 major groups (metabolism, cell signaling, transcription, transport physiology, growth, and differentiation, immune system, cell adhesion, house keeping, cell cycle, translation, apoptosis, and unknown genes (ESTs)] (Figure 1). We found that genes with unknown functions represented the major portion of genes with altered expression levels in the HBV immune twin.
We are interested in the immune system-related genes that are putative candidates for conferring HBV immunity. Among the up- or down-regulated genes, eight genes were related to immune responses including several cytokine/chemokine signaling-related genes (Table 1). Three genes, namely cytokine PBEF, interleukin IL-18 receptor (R) accessory protein and TNF-αIP1 were upregulated in the immunity twin. PBEF is a cytokine transcribed in human bone marrow, liver tissue and muscle[9,10] and synergized with stem cell factor (SCF) and IL-7 activating pre-B-cell colony formation[11]. IL-18R accessory protein enhances the IL-18 binding activity of IL-18R1 and is required for the activation of nuclear factor-kappa B (NF-ĸB) and mitogen-activated protein kinase 8 (MAPK 8) in response to IL-18. TNF-αIP1 is first identified as a primary response gene in human umbilical vein endothelial cells towards TNF-α stimulation[12], but yet with no known function.
| UniGene ID | Gene name | Known function(s) | Fold change |
| Hs.244613 | Signal transducer and activator of transcription 5b (STAT 5b) | Signal transducer of IL-2, IL-4, CSF1, and different growth hormones. I mportant for TCR signaling, apoptosis, adult mammary gland development, and sexual dimorphism of liver gene expression | –4.2 |
| Hs.239138 | Pre-B-cell colony-enhancing factor (PBEF) | A cytokine that increases the expression of IL-6 and IL-8 in fetal membrane and may be important in both normal spontaneous labor and infection-induced preterm labor | 2.2 |
| Hs.158315 | Interleukin 18 receptor accessory protein (IL-18RAP) | An accessory subunit of the heterodimeric receptor for IL-18 enhances the IL-18 binding a ligand binding subunit of IL-18 receptor. The coexpression of IL-18R1 and this protein is required for the activation of NF-κB and MAPK8 (JNK) in response to IL-18 | 3.0 |
| Hs.76090 | Tumor necrosis factor-alpha-induced protein 1 (endothelial) (TNF-αIP1) | Unknown | 2.3 |
| Hs.225948 | Chemokine (C-C motif) ligand 27 (CCL27) | Cytokine that plays a role in mediatinghoming of lymphocytes to cutaneous sites. It specifically binds to chemokine receptor 10 (CCR10). Studies of murine protein indicate that these protein-receptor interactions have a pivotal role in T-cell-mediated skin inflammation | -2.5 |
| Hs.301921 | C-C chemokine receptor type 1 (C-C CKR-1) | Cytokine receptor important for host protection from inflammatory response and susceptibility to virus and parasite | -2.7 |
| Hs.57735 | Scavenger receptor class F, member 1 (SCARF1) | Scavenger receptor that has roles in the binding and degradation of acetylated low density lipoprotein and may be involved in atherosclerosis | -2.2 |
| Hs.4930 | Low density lipoprotein receptor-related protein 4 (LRP4) | A membrane protein which may be involved in calcium ion binding | -2.8 |
Five genes were downregulated in the HBV-immuned twins. They are the signal transducer and activator of transcription 5b (Stat5b), which is crucial for normal immune function and T-cell-mediated mitogenic signals and is a key signal transducer of T-cell receptor[13]. The chemokine (C-C motif) ligand 27 (CCL27) and C-C chemokine receptor type 1 (C-C CKR-1) are important for host inflammatory response. We also observed the downregulation of the scavenger receptor class F member 1 (SCARF1) and low-density lipoprotein receptor-related protein 4. Scavenger receptors encompass a broad range of molecules involved in receptor-mediated endocytosis of selected polyanionic ligands, including modified low-density lipoproteins (LDL)[14]. Further investigating the role of these genes and the molecular differences between immune siblings and HBV chronic carrier siblings may help us in designing therapeutic measures that modulate the immune system of patients in controlling HBV infection.
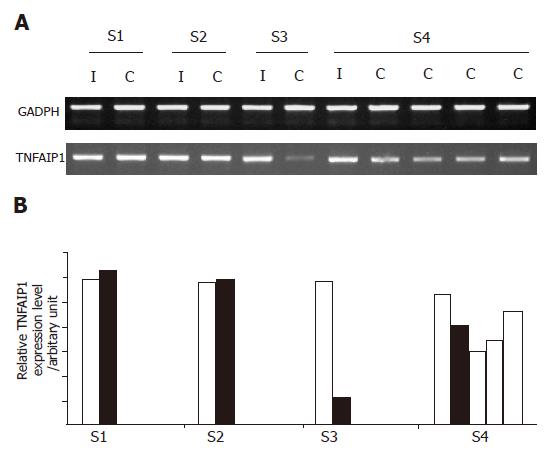
Of the eight immune system-related genes, TNF-αIP1 demonstrated the highest frequency of differential expression among the other groups of HBV-infected HLA-matched siblings with different disease outcomes as determined by semi-quantitative RT-PCR (Figure 2A). As shown in Figure 2B, the expression level of TNF-αIP1 was lower in HBV carriers than in their HBV immune siblings in two siblings groups. Noticeably, the expression of TNF-αIP1 was significantly downregulated in HBV carrier siblings of group 3 and all four carrier siblings displayed down-regulation of TNF-αIP1. Suggesting that TNF-αIP1 may be a gene differentially expressed in PBMCs of HBV immune and carrier patients.
Recent studies suggest that cytokine response plays an important role in successful host defense against HBV infection. In transgenic mouse and chimpanzee model, cytotoxic T lymphocytes do not directly kill HBV-infected hepatocytes, but inhibit the viral replication through the actions of TNF-α and interferon (IFN-γ)[15,16]. Romero and Lavine[17] demonstrated that TNF-α and IFNs can downregulate the activity of HBV core/pregenomic (C/P) promoter, hence contributing to viral clearance. Moreover, activated intrahepatic antigen-presenting cells can inhibit liver HBV replication by secreting IL-12 and TNF-α[18]. One of the downstream mechanisms of how TNF-α modulates HBV infection is via the activation of NF-ĸB, which can inhibit HBV replication[19]. Interestingly, we have shown that TNF-αIP1 (a TNF-α inducible gene) expresses at a higher level in HBV immunized twin. TNF-αIP1 was first identified as an endothelial primary response gene towards TNF-α (then named B12), which induces TNF-αIP1 expression rapidly and transiently. Biochemical characterization suggests that TNF-αIP1 is a 36 ku protein, which may play a regulatory role and locate intracellularly[12]. The role of TNF-αIP1 in TNF-α-mediated NF-κB activation and HBV replication inhibition is of interest for further study.
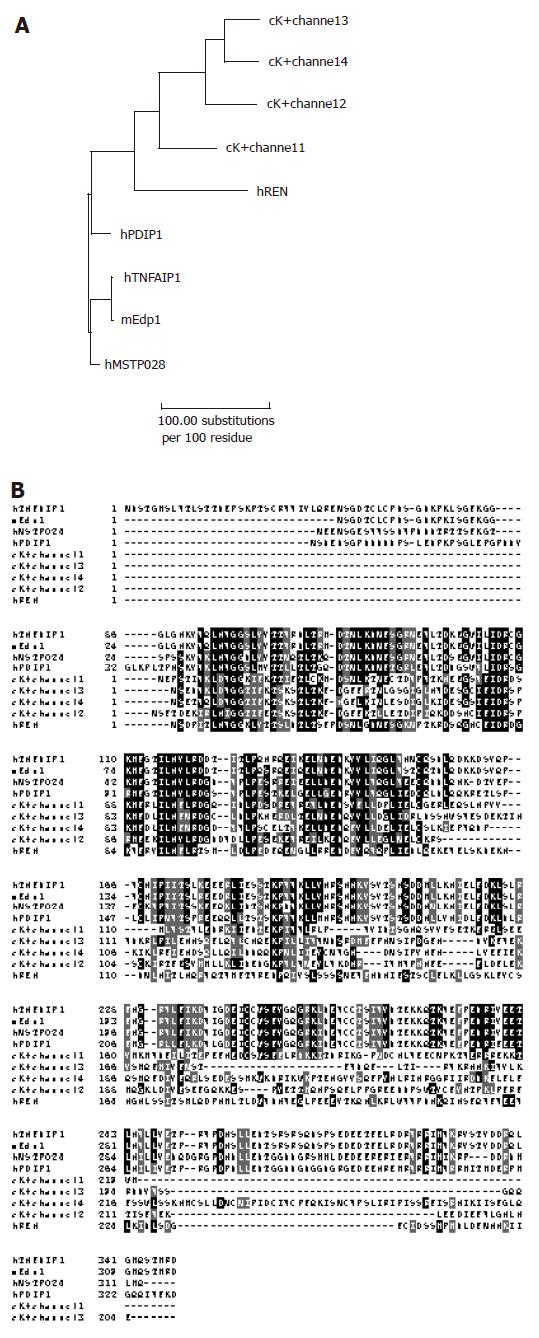
Phylogenetic analysis showed that TNF-αIP1 could evolve into human and mouse endothelial protein 1 (Edp1), reflecting the fact that Edp1 is likely the mouse ortholog of TNF-αIP1 (Figure 3A). Predicted from the amino acid sequence of TNF-αIP1 (A41784), there is a K+ channel tetramerization domain in the middle of the sequence. This N-terminal, cytoplasmic tetramerization domain (T1) of voltage-gated K+ channels encodes molecular determinants for subfamily-specific assembly of alpha-subunits into functional tetrameric channels[20]. It is distantly related to the BTB/POZ domain which plays an oligomerization role in the function of this protein. Interestingly, the K+ channel tetramerization domain is the common domain feature shared by many TNF-α inducible proteins. For instance, mouse TNF-α inducible proteins such as Edp1 share high homology with the entire amino acid sequence of TNF-αIP1 (97% identity) (Figures 3A and B). It is also noticed that there was a group of C elegan proteins containing the K+ channel tetramerization domain. However, the function of this group of proteins is unknown.
Among the proteins containing K+ channel tetramerization domain, polymerase delta-interacting protein 1 (PDIP1) is best characterized[21]. PDIP1 is a TNF-α inducible protein and plays a role in linking cytokine activation and DNA replication in the liver as well as in other tissues. It localizes inside the nuclei and interacts with DNA polymerase δ small subunit (p50) and proliferating cell nuclear antigen (PCNA). In addition, it modulates DNA polymerase δ activity in a PCNA dependent manner. Importantly, it shares high homology (62% identity) with TNF-αIP1 (B12). PDIP1 is involved in TNF-α-mediated hepatocyte regeneration and stimulates hepatocyte proliferation upon TNF-α signal[21]. It is of interest to know whether this is also true for TNF-αIP1 using cell proliferation assay.
| 1. | Yeoh EK, Chang WK, Kwan JPW. Epidemiology of viral hepatitis B infection in Hong Kong. In Lam SK, Lai CL, Yeoh EK (editors): Viral hepatitis B infection: Vaccine and control. Singapore: World Scientific 1984; 33-41. |
| 2. | Lok AS, Lai CL, Wu PC, Wong VC, Yeoh EK, Lin HJ. Hepatitis B virus infection in Chinese families in Hong Kong. Am J Epidemiol. 1987;126:492-499. [PubMed] |
| 3. | Stevens CE, Beasley RP, Tsui J, Lee WC. Vertical transmission of hepatitis B antigen in Taiwan. N Engl J Med. 1975;292:771-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 759] [Cited by in RCA: 702] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 4. | Lok AS, Liang RH, Chung HT. Recovery from chronic hepatitis B. Ann Intern Med. 1992;116:957-958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 5. | Lau GK, Lok AS, Liang RH, Lai CL, Chiu EK, Lau YL, Lam SK. Clearance of hepatitis B surface antigen after bone marrow transplantation: role of adoptive immunity transfer. Hepatology. 1997;25:1497-1501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 149] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 6. | Lau GK, Liang R, Lee CK, Yuen ST, Hou J, Lim WL, Williams R. Clearance of persistent hepatitis B virus infection in Chinese bone marrow transplant recipients whose donors were anti-hepatitis B core- and anti-hepatitis B surface antibody-positive. J Infect Dis. 1998;178:1585-1591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 45] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 7. | Lau GK, Lie AK, Kwong YL, Lee CK, Hou J, Lau YL, Lim WL, Liang R. A case-controlled study on the use of HBsAg-positive donors for allogeneic hematopoietic cell transplantation. Blood. 2000;96:452-458. [PubMed] |
| 8. | Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, Xiao H, Rogers KE, Wan JS, Jackson MR. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med. 1999;5:117-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 507] [Cited by in RCA: 494] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 9. | Samal B, Sun Y, Stearns G, Xie C, Suggs S, McNiece I. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol Cell Biol. 1994;14:1431-1437. [PubMed] |
| 10. | Ognjanovic S, Bryant-Greenwood GD. Pre-B-cell colony-enhancing factor, a novel cytokine of human fetal membranes. Am J Obstet Gynecol. 2002;187:1051-1058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 150] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 11. | Rongvaux A, Shea RJ, Mulks MH, Gigot D, Urbain J, Leo O, Andris F. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur J Immunol. 2002;32:3225-3234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 12. | Wolf FW, Marks RM, Sarma V, Byers MG, Katz RW, Shows TB, Dixit VM. Characterization of a novel tumor necrosis factor-alpha-induced endothelial primary response gene. J Biol Chem. 1992;267:1317-1326. [PubMed] |
| 13. | Welte T, Leitenberg D, Dittel BN, al-Ramadi BK, Xie B, Chin YE, Janeway CA, Bothwell AL, Bottomly K, Fu XY. STAT5 interaction with the T cell receptor complex and stimulation of T cell proliferation. Science. 1999;283:222-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 108] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 14. | Peiser L, Mukhopadhyay S, Gordon S. Scavenger receptors in innate immunity. Curr Opin Immunol. 2002;14:123-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 337] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 15. | Guidotti LG, Ishikawa T, Hobbs MV, Matzke B, Schreiber R, Chisari FV. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity. 1996;4:25-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 840] [Cited by in RCA: 842] [Article Influence: 28.1] [Reference Citation Analysis (1)] |
| 16. | Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825-829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 949] [Cited by in RCA: 920] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 17. | Romero R, Lavine JE. Cytokine inhibition of the hepatitis B virus core promoter. Hepatology. 1996;23:17-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 72] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 18. | Kimura K, Kakimi K, Wieland S, Guidotti LG, Chisari FV. Activated intrahepatic antigen-presenting cells inhibit hepatitis B virus replication in the liver of transgenic mice. J Immunol. 2002;169:5188-5195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 88] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Biermer M, Puro R, Schneider RJ. Tumor necrosis factor alpha inhibition of hepatitis B virus replication involves disruption of capsid Integrity through activation of NF-kappaB. J Virol. 2003;77:4033-4042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 141] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 20. | Bixby KA, Nanao MH, Shen NV, Kreusch A, Bellamy H, Pfaffinger PJ, Choe S. Zn2+-binding and molecular determinants of tetramerization in voltage-gated K+ channels. Nat Struct Biol. 1999;6:38-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 119] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 21. | He H, Tan CK, Downey KM, So AG. A tumor necrosis factor alpha- and interleukin 6-inducible protein that interacts with the small subunit of DNA polymerase delta and proliferating cell nuclear antigen. Proc Natl Acad Sci USA. 2001;98:11979-11984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
Science Editors Wang XL and Guo SY Language Editor Elsevier HK