Published online Dec 7, 2005. doi: 10.3748/wjg.v11.i45.7097
Revised: June 6, 2005
Accepted: June 11, 2005
Published online: December 7, 2005
AIM: To study the secretory expression of human hepatocyte growth factor (hdHGF) gene in Pichia pastoris.
METHODS: The full-length gene of human cDNA encoding the deleted variant of hdHGF was cloned by RT-PCR and overlapping-fragment PCR technique using mRNA of human placenta as a template. The cloned hdHGF cDNA was inserted into the Escherichia coli-yeast shuttle vector of pPIC9. The constructed plasmid, pPIC9-hdHGF, was transformed into the GS115 cells of the methylotrophic yeast, P pastoris, using a chemical method. The Mut+ transformants were screened to obtain high-expression strains by the test and analysis of expressed products of shake-flask culture. A secretory form of rhdHGF was made with the aid of the leader peptide sequence of Saccharomyces cerevisiae α-factor.
RESULTS: The expressed products, which showed a band of molecular mass of about 80 ku, were observed on 15% SDS-PAGE and identified by Western blotting and N-terminal amino acid sequencing. In the high cell density culture of 5 L fermentor by fed-batch culture protocol, the cell biomass was reached at approximately 135 g (DCW)/L. The productivity of secreted total supernant protein concentration attained a high-level expression of more than 8.0 g/L and the ratio of rhdHGF band area was about 12.3% of the total band area scanned by SDS-PAGE analysis, which estimated that the product of rhdHGF was 500-900 mg/L.
CONCLUSION: The P pastoris system represents an attractive tool of generating large quantities of hdHGF for both research and industrial purposes.
- Citation: Liu ZM, Zhao HL, Xue C, Deng BB, Zhang W, Xiong XH, Yang BF, Yao XQ. Secretory expression and characterization of a recombinant-deleted variant of human hepatocyte growth factor in Pichia pastoris. World J Gastroenterol 2005; 11(45): 7097-7103
- URL: https://www.wjgnet.com/1007-9327/full/v11/i45/7097.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i45.7097
Hepatocyte growth factor (HGF) was identified initially as a mitogen for hepatocytes, called as scatter factor (SF) and fibroblast-derived tumor cytotoxic factor (F-TCF) as well as fibroblast-derived growth factor called plasminogen-like growth factor (PLGF)[1-3]. Nakamura et al[16]. reported that HGF could be purified from the serum of partially hepatectomized rats. Subsequently, HGF has been purified from rat platelets and its subunit structure is determined. The purification of human HGF from human plasma is described by Godowski et al[5].
The gene locus of human HGF is assigned to chromosome 7q21.1. The genomic gene consists of 18 exons and 17 introns, and spans about 70 kb. The whole length form of human hepatocyte growth factor (preproHGF) consists of 727 amino acids and the mature form of hHGF is composed of 674 amino acids, corresponding to the major form purified from human serum[5]. HGF is a disulfide-linked heterodimer derived by proteolytic cleavage of the human pro-hormone between amino acids R494 and V495. This cleavage process generates a molecule composed of an alpha-subunit of 440 amino acids (MW 69 ku) and a beta-subunit of 234 amino acids (MW 34 ku). The nucleotide sequence of hHGF cDNA reveals that both the alpha-and the beta-chains are contained in a single open reading frame coding for a pre-pro precursor protein. In the predicted primary structure of mature hHGF, an interchain S-S bridge is formed between Cys 487 of the alpha-chain and Cys 604 in the beta-chain. The N-terminus of the alpha-chain is preceded by 54 amino acids, starting with a methionine group. This segment includes a characteristic hydrophobic leader (signal) sequence of 31 residues and the prosequence. The alpha-chain starts at amino acid (aa) 55 and contains four Kringle domains. The Kringle 1 domain extends from about aa 128 to about aa 206, the Kringle 2 domain is between about aa 211 and about aa 288, the Kringle 3 domain extends from about aa 303 to about aa 383, the Kringle 4 domain extends from about aa 391 to about aa 464 of the alpha-chain. It will be understood that the definition of the various Kringle domains is based on their homology with Kringle-like domains of other proteins (prothrombin, plasminogen). Therefore, the above limits are only approximate. Until now, the function of these Kringles has not been determined. The beta-chain of hHGF shows high homology to the catalytic domain of serine proteases (38% homology to the plasminogen serine protease domain). However, two of the three residues, which form the catalytic triad of serine proteases, are not conserved in hHGF. Therefore, despite its serine protease-like domain, hHGF appears to have no proteolytic activity and the precise role of the beta-chain remains unknown. HGF contains four putative glycosylation sites, which are located at positions 294 and 402 of the alpha-chain and at positions 566 and 653 of the beta-chain. Wild-type human HGF gene in vivo exists in the polymorphism. It has been observed that human HGF has a few natural variants. For example, hdHGF-encoded HGF molecule lacking five amino acids in the Kringle 1 domain (FLPSS) is fully functional[6-8].
HGF biological activity refers to any mitogenic, motogenic or morphogenic activities exhibited by wild-type human HGF or hdHGF, which have a broad spectrum of mitogenic cell specificity that can promote the proliferation of hepatocytes, endothelial cells, fibroblasts, melanocytes, and epithelial cells etc.[1,4], inhibit the growth of some tumor cell lines such as HepG2, B6/F1, and KB from tumorigenic target cell lines[9]. Recent studies displayed that hdHGF can exert many important biological effects mediated via their specific tyrosine kinase receptor, C-met. Even to the extent, hdHGF has more significant biological effects on promoting the regeneration of hepatocytes and kidney epithelial cells compared to wild-type human HGF, suggesting that hdHGF has the therapeutic effect in vivo on liver injury[2].
Recently, methylotrophic yeast Pichia pastoris has become a dominant tool in molecular biology for the production of recombinant proteins. P pastoris is known for its high-level expression of heterologous proteins and its tightly regulated alcohol oxidase 1 (AOX1) gene promoter[10]. P pastoris can be easily grown to high cell densities using defined minimal media and is able to introduce eukaryotic post-translational modifications[11,12]. The techniques for molecular genetic manipulation are similar to those well established for Saccharomyces cerevisiae. At present, P pastoris as an efficient protein expression system can be fermented routinely in large scale to meet the industrial demands of interest proteins[13-15]. In the present report, we have described the recombinant production of hdHGF in P pastoris and its characterization.
P pastoris host strain GS115 (His+Mut+) and secretion expression vector pPIC9 were purchased from Invitrogen (San Diego, CA, USA). E coli DH5α was used for routine plasmid amplification and the cloned vector of pUC19 was maintained in our laboratory. SuperscriptTM II RNase H-Reverse transcriptase was purchased from GibcoBRL. Human placenta mRNA was obtained from Clontech Co. Expand™ High Fidelity PCR System was purchased from Boehringer Mannheim Co. Yeast nitrogen base, D-biotin, yeast extract and tryptone were obtained from Sangon (Shanghai, China). EcoRI, NotI, SalI, XbaI, SphI, T4 DNA ligase, and Taq DNA polymerase were obtained from TaKaRa Biotechnology (Dalian, China). Anti-hHGF antibody was purchased from Santa Cruz Biotechnology Co.
The whole length gene of human cDNA encoding the deleted variant of hdHGF was amplified by RT-PCR and overlapping fragments were amplified by PCR technique using mRNA of human placenta as the template. Three pairs of PCR primers for amplified hdHGF fragments were designed as follows.
In primer M1, single bottom line stands for SphI and two lines for SalI. Oblique boldface capital letters (included 8 codons) represent the frequently used codons in the highly expressed P pastoris genes. In primer M2, single bottom line stands for XbaI and two lines for NotI. Its complementary chain encodes the sequence for TACAAGGTTCCACAGTCTTAA (included 6 codons) and the oblique boldface capital letters represent the bias of codons in the highly expressed P pastoris genes. Therefore, the codons encoding N-and C-terminal amino acids of the deleted variant of hdHGF were amplified by RT-PCR and overlapping-fragment PCR technique, using mRNA of human placenta as the template, which has the advantage to acquire high-level expression of foreign genes in P pastoris.
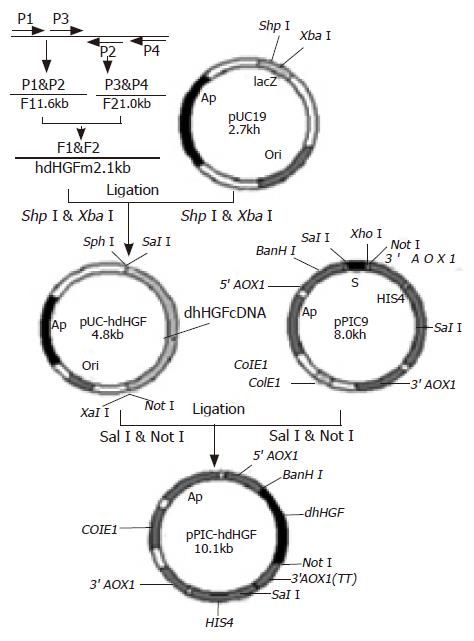
A forward primer(P1: 5’TTCTTTCACCCAGGCATCTC3’) and a reverse primer (P2: 5’CTATGTTTGTTCGTGTTGGAATCC3’) as well as another forward primer (P3: 5’GTGGGACAAAGAACATGGAAGACTTAC3’) and its reverse primer (P4: 5’GCTTCAGACACACTTACTT CAGCTA3’) were designed to synthesize the two cDNA fragments based on the hdHGF sequence reported by Nakamura et al[16], namely one fragment (F1, about 1.6 kb) was amplified using a pair of primers P1 and P2 and the other fragment (F2, approximately 1.0 kb) was amplified using a pair of primers P3 and P4. Overlapping-fragment amplification using F1 and F2 fragments as templates was performed by routine PCR procedure using a pair of primers M1 and M2 (Figure 1). The cDNA product obtained from RT-PCR was modified by introducing SphI and SalI sites at the 5’end and XbaI and NotI sites as well as a TAA stop codon at the 3’end. Thirty-five cycles of PCR were performed: denaturation at 94 °C for 60 s, annealing at 55 °C for 60 s, extension at 72 °C for 90 s, and then a further extension at 72 °C for 10 min. The PCR procedures were carried out according to the standard procedures published earlier[17].
The PCR products were digested with SphI and XbaI, and cloned into the same enzyme digested vector pUC19. The recombinant vectors were transformed into E coli DH5α. The recombinant transformants were acquired via the blue-white colony screening in the agar medium containing X-gal and characterized using restriction endonucleases SphI and XbaI. The gene sequence analysis of the recombinant pUC-hdHGF was carried out by Sanger’s dideoxynucleotide DNA sequencing. The verified hdHGF cDNA fragment with SalI and NotI was cloned into the site of expression vector pPIC9 digested with XhoI and NotI enzymes. The recombinant plasmids were transformed into E coli strains of JM109. Screening and selection of expression plasmid clones containing hdHGF cDNA fragments through the identification with restriction endonucleases BamHI and NotI, resulted in the plasmid pPIC9-hdHGF containing hdHGF gene under the control of AOX1 promoter and in-frame with α-factor signal sequence (Figure 2).
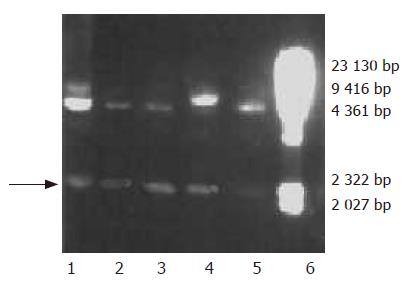
Plasmids used for transformation were linearized with SalI. The SalI-linearized pPIC9-hdHGF or parent pPIC9 was transformed into his4 competent P pastoris GS115 cells by a chemical method. After the growth on minimal dextrose medium (MD) plates at 30 °C for 3 d, several colonies containing the linearized pPIC9-hdHGF fragment were selected for PCR confirmation by colony PCR, which was designed to amplify the 200-bp special sequence of pPIC9-hdHGF using a pair of primers, namely P5 (sense, 5’GTGGGACAAGAACATGGAAGA CTTA3’) and P6 (antisense, 5’CTATGTTTGTTCGTGTTGGAATCC3’).
Ten colonies were used to inoculate 10 mL buffered minimal glycerol-complex medium (BMGY) in a 50 mL shake flask, respectively. After being shook at 250 r/min for 2 d at 30 °C, the cells were pelleted and resuspended in a 2 mL buffered minimal methanol-complex medium (BMMY). Following the additional 2 d of induction at 30 °C, the samples of expressed hdHGF in culture supernatants were determined and hdHGF in culture supernatants was also analyzed by SDS-PAGE.
The clones exhibiting the highest level expression of hdHGF were selected for fed-batch fermentations which were carried out in a 5-L working volume bioreactor using a BIOFLO 3000 (New Brunswick Scientific) interfaced with AFS-Biocommand Bioprocessing software version 2.6 (New Brunswick Scientific) for data acquisition and supervisory control.
Seed culture for the bioreactor was started from the fresh glycerol stock and inoculated directly into 500-mL shake flasks (50-mL working volume) containing a minimum glycerol medium (1.34% YNB, 1% glycerol, and 1.61 μmol/L biotin). After 24 h of growth, seed culture was inoculated with 1% inoculum. After 16-20 h of growth, seed culture was used to inoculate the bioreactor. Ten percentage of the inoculum was used for the inoculation of a 5-L bioreactor containing 2.-L medium of high-cell density fermentation, comprising of 10× basal salts (42 mL/L 85% H3PO4, 1.8 g/L CaSO4∙2H2O, 28.6 g/L K2SO4, 50 g/L glycerol, 23.4 g/L MgSO4∙7H2O, 6.5 g/L KOH, and 4.35 mL/L 10∙PTM1 salts, 6.0 g/L CuSO4· 5H2O, 0.08 g/L KI, 3.0 g/L MnSO4∙H2O, 20.0 g/L ZnCl2, 0.02 g/L HBO4, 65 g/L FeSO4∙7H2O, 0.2 g/L Na2MoO4∙2H2O, 0.5 g/L CoCl2, 0.2 g/L biotin, and 5 mL/L H2SO4, buffered to pH 5.5 using 2 mol/L NH4OH). Dissolved oxygen was maintained at over 20% air saturation at 30 °C and aeration was maintained at 2 vvm. pH was maintained at 5.5 and fermentation was carried out in two phases. Growth phase consisted of a glycerol batch phase and cells were grown batch-wise until glycerol in the medium was utilized. To achieve a high cell density, the glycerol (50% glycerol, 4.3 mL/L PTM1, feeding rate: 18 mL/h∙L) fed-batch phase was initiated and lasted for 6-10 h. Production phase consisted of a methanol fed-batch phase when cells were induced by methanol (100% methanol plus 12 mL/L PTM1 salts). The methanol feed rate was gradually increased over a period of 6 h-6 mL/h and the fermentation continued for an additional 46-92 h. Expressed hdHGF in culture supernatants was analyzed by SDS-PAGE and the concentration of secreted total supernant proteins was also determined at different intervals of induction phase using the standard curve analysis of human serum albumin (HSA).
Based on the hdHGF sequence reported by Nakamura et al[16], we designed three pairs of PCR primers for amplified hdHGF fragments. A forward primer P1 and a reverse primer P2 and another forward primer P3 and its reverse primer P4 were designed to synthesize the two fragments. The two fragments of hdHGF PCR products about 1 570 (F1) and 970 bp (F2), respectively, were also clearly seen in 1% agarose gel electrophoresis stained with 5 mg/mL ethidium bromide amplified with P1-P2 and P3-P4 primer pairs (Figure 3A). Overlapping-fragment PCR amplification using F1 and F2 fragments as templates was performed by routine PCR procedure using a pair of primers M1 and M2 and the PCR product of full-length gene of hdHGF was revealed (Figure 3B).
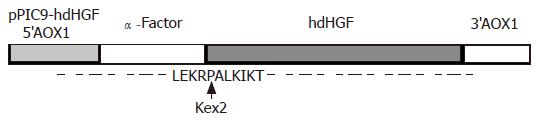
A cDNA encoding the mature form of hdHGF was used in these experiments. This cDNA consisted of 2.1 kb (Figure 2B) with an open-reading frame encoding a 669 amino acid peptide. The DNA fragment encoding the mature hdHGF was digested with XhoI and NotI from pUC19 vector and cloned into the same enzyme digested vector pPIC9 downstream of the alcohol oxidase I (AOX I) promoter (Figures 1 and 3). The resultant construct harbored a single open reading frame encoding a 85 amino acid translation product consisting of the α-factor secretion leader peptide (Figures 1 and 3). Prior to the secretion of the peptide into the culture medium, the signal peptide should be cleaved off by the KEX2 gene products at the site (Glu-Lys-Arg-X) (Figure 1). The integrity of the recombinant plasmid was confirmed by direct DNA sequencing. Enzyme identification of the recombinant plasmids of pPIC9-hdHGF digested by BamHI and NotI is shown in Figure 4. This constructed vector was linearized with SalI and transformed into the competent cells of P pastoris GS115. The transformants were selected on MD plates and confirmed by colony PCR. Forty-one colonies presenting strong amplification products were used for small-scale expression trials and the amount of the recombinant peptide was determined by SDS-PAGE.
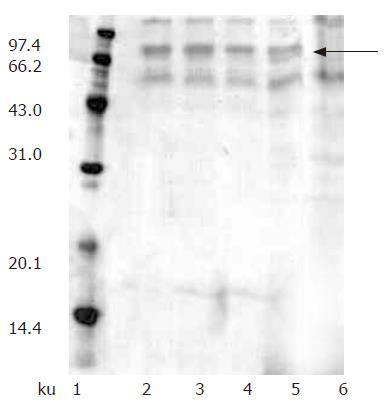
We investigated the expression of recombinant hdHGF by both Muts and Mut+ (GS115) strains in shake flasks. Since SDS-PAGE analysis revealed that the hdHGF level expressed by Muts strain was much higher than that of Mut+ (data not shown), Muts was chosen for the expression of th e growth factor. In addition, Muts phenotype was selected over the Mut+ phenotype because of the latter’s higher oxygen requirement that could result in oxygen-deficient conditions within the bioreactor. Fifty transformants (Muts) were used for the expression studies in shake flask experiments and secretion of the recombinant hdHGF into the culture medium was determined by SDS-PAGE analysis. The expression experiments were performed to screen out four high-level expression strains of hdHGF, which were named as HG209, HG211, HG305, and HG309 (Figure 5).
The selected clones with the highest expression level were chosen for fed-batch cultivation. A time-course study of secretion of hdHGF revealed a gradual accumulation of recombinant cytokines. The effect of induction pH on the production of recombinant cytokines was investigated. P pastoris is known to grow over a wide pH range from 3 to 7, with a minimal effect of pH on the growth rate. However, pH could significantly affect the productivity of secreted recombinant proteins in the fermentation broth. To find out the optimal pH for the expression of recombinant hdHGF, we conducted experiments with pH between 3.5 and 6.5 during the fed-batch production phase. The highest yield of recombinant hdHGF was observed at pH 5.5 (Figure 6).
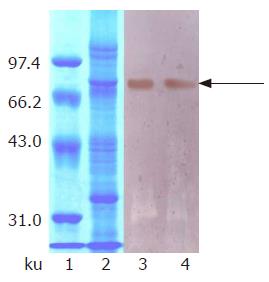
Protein expression was initiated by changing carbon source from glycerol to methanol. At first, we attempted to express hdHGF in baffled shake flasks and obtained about 50 mg/L of hdHGF secreted into the medium after 72-h induction. For more systematic production of hdHGF, we expressed the protein using fermenter cultures. Figure 6 shows the secretion level of hdHGF using 5 L fermenter. Upon depletion of glycerol, the dry cell weight reached 45.6 g/L. A glycerol fed-batch phase was performed for an additional 6-12 h and the cell biomass reached at approximately 135 g (DCW)/L. Induction of hdHGF was initiated by the addition of 100% methanol containing 12 mL of PTM1 trace salts/L. Sample analysis at different intervals was also performed to show the increasing amounts of recombinant hdHGF presented in the culture medium with increasing induction time until 96 h. Dissolved oxygen (DO) was monitored by DO sensor and the oxygen transfer rate 1 min after turning off the carbon source feed. Dissolved oxygen was maintained between 20% and 30% air saturation in the two-phase fermentation. The secreted total supernant protein concentration in the induction phase was traced and observed to attain the high-level expression of more than 8.0 g/L after 84-96 h of induction cultivation, which was determined using the standard curve analysis of human serum albumin (HSA). The scanning result showed that the secreted rhdHGF protein band (in lane 3 of Figure 6) achieved about 12.3% of the total supernatant proteins. By comparison with the standard protein markers, the estimated product of rhdHGF was 500-900 mg/L. The maximum secretion yield was approximately 980 mg/L (Figure 6). Upon induction by methanol, four clones secreted a specific 80-ku protein with the same size as the standard HGF. The productivity varied among the four high-level expression strains of hdHGF.
These clones indicated that the highest product of rhdHGF was named HG209 (GS115/pPIC9-hdHGF) and selected for further analysis. Western blotting analysis showed that the 80-ku protein band from HG209 (GS115/pPIC9-hdHGF) reacted specifically with the rabbit anti-hHGF antiserum (Figure 6). The N-terminal sequence of the recombinant hdHGF was determined to be PALKI, which was identical to the N-terminal sequence of native hdHGF (Figure 1).
hdHGF is a large complex protein comprising of 669 amino acids. It is the most potent multifunctional cytokine on the regeneration of hepatocytes and kidney epithelial cells compared to wild-type human HGF[20]. It can promote cell division, migration, and differentiation. Its receptor is the product of oncogene c-met. Besides being a nutritional factor of liver and kidney, hdHGF also promotes angiogenesis for peripheral artery disease and myocardial ischemia[20] and can affect synthesis of extracellular matrix and matrix metalloproteinases and tissue inhibitor of metalloproteinases in autosomal dominant polycystic kidney disease cyst-lining epithelial cells[21].
Since the concentration of native HGF in plasma is very low, purification of HGF from plasma is very difficult. It was reported that HGF is expressed in foreign gene expression systems such as mammalian cells, CHO cells[22,23], insect cells[24], and gene therapy[25]. Dang et al[26] and Li et al[27]. reported that the HGF gene is expressed in E coli and P pastoris. However, the expression of recombinant-deleted variants of human hepatocyte growth factor (hdHGF) has not been reported in yeast expression system. Therefore, in this investigation, we used the methylotrophic yeast P pastoris as the host for the high-level expression and secretion of recombinant hdHGF. Recombinant hdHGF was successfully secreted by P pastoris and the productivity of secreted total supernatant protein concentration attained high-level expression of more than 8.0 g/L and the ratio of rhdHGF band area was about 12.3% of the total band area scanned by SDS-PAGE analysis, which estimated the product of rhdHGF to be 500-900 mg/L. It had an approximately fivefold increase in productivity compared to that of HGF expressed in P pastoris[27]. Western blot analysis showed that the 80-ku protein band of GS115 (pPIC9-hdHGF) reacted specifically with the rabbit anti-hHGF antibody. N-terminal sequencing revealed that recombinant rhdHGF had the correct N-terminal amino acid sequence. These results suggest that the optimization of bias codons of P pastoris encoding N-and C-terminal amino acids of hdHGF via PCR-mediated codon replacement can acquire high-level expression of foreign genes in P pastoris. Nakamura et al[16]. reported that native hdHGF is composed of an alpha-subunit of 440 amino acids (MW 69 ku) and a beta-subunit of 234 amino acids (MW 34 ku). However, we found that the recombinant hdHGF produced by P pastoris was translated as a single-chain polypeptide comprising of 669aa, which was not cut by the proteolytic cleavage of the human pro-hormone between amino acids R494 and V495 in host cells of P pastoris.
In conclusion, though further characterization, bioassay, and optimization of the expression and cultivation of recombinant hdHGF by P pastoris are required, this expression system of hdHGF is expected to be a powerful tool in the industrial production of this foreign protein.
| 1. | Kataoka H, Miyata S, Uchinokura S, Itoh H. Roles of hepatocyte growth factor (HGF) activator and HGF activator inhibitor in the pericellular activation of HGF/scatter factor. Cancer Metastasis Rev. 2003;22:223-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 101] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 2. | Kinosaki M, Yamaguchi K, Murakami A, Morinaga T, Ueda M, Higashio K. Analysis of deleted variant of hepatocyte growth factor by alanine scanning mutagenesis: identification of residues essential for its biological function and generation of mutants with enhanced mitogenic activity on rat hepatocytes. FEBS Lett. 1998;434:165-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 3. | Weidner KM, Arakaki N, Hartmann G, Vandekerckhove J, Weingart S, Rieder H, Fonatsch C, Tsubouchi H, Hishida T, Daikuhara Y. Evidence for the identity of human scatter factor and human hepatocyte growth factor. Proc Natl Acad Sci U S A. 1991;88:7001-7005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 460] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 4. | Matsumoto K, Nakamura T. Emerging multipotent aspects of hepatocyte growth factor. J Biochem. 1996;119:591-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 520] [Cited by in RCA: 501] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 5. | Godowski PJ, Lokker NA, Mark MR. Single-chain hepatocyte growth factor variants. United States Patent 5580963. 1996;. |
| 6. | Shima N, Tsuda E, Goto M, Yano K, Hayasaka H, Ueda M, Higashio K. Hepatocyte growth factor and its variant with a deletion of five amino acids are distinguishable in their biological activity and tertiary structure. Biochem Biophys Res Commun. 1994;200:808-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 69] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 7. | Kinosaki M, Yamaguchi K, Murakami A, Ueda M, Morinaga T, Higashio K. Identification of heparin-binding stretches of a naturally occurring deleted variant of hepatocyte growth factor (dHGF). Biochim Biophys Acta. 1998;1384:93-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 8. | Yasuda H, Imai E, Shiota A, Fujise N, Morinaga T, Higashio K. Antifibrogenic effect of a deletion variant of hepatocyte growth factor on liver fibrosis in rats. Hepatology. 1996;24:636-642. [PubMed] [DOI] [Full Text] |
| 9. | Shiota G, Rhoads DB, Wang TC, Nakamura T, Schmidt EV. Hepatocyte growth factor inhibits growth of hepatocellular carcinoma cells. Proc Natl Acad Sci USA. 1992;89:373-377. [PubMed] |
| 10. | Cregg JM, Vedvick TS, Raschke WC. Recent advances in the expression of foreign genes in Pichia pastoris. Biotechnology (N Y). 1993;11:905-910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 446] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 11. | Brierley RA, Bussineau C, Kosson R, Melton A, Siegel RS. Fermentation development of recombinant Pichia pastoris expressing the heterologous gene: bovine lysozyme. Ann N Y Acad Sci. 1990;589:350-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 104] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 12. | O'Leary JM, Radcliffe CM, Willis AC, Dwek RA, Rudd PM, Downing AK. Identification and removal of O-linked and non-covalently linked sugars from recombinant protein produced using Pichia pastoris. Protein Expr Purif. 2004;38:217-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 13. | Zhang W, Li ZJ, and Foster A Agblevor. Microbubble fermentation of recombinant Pichia pastoris for human serum albumin production. Process Biochemistry. 2005;40:2073-2078. [RCA] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 14. | Werten MW, van den Bosch TJ, Wind RD, Mooibroek H, de Wolf FA. High-yield secretion of recombinant gelatins by Pichia pastoris. Yeast. 1999;15:1087-1096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 15. | Cregg JM, Cereghino JL, Shi J, Higgins DR. Recombinant protein expression in Pichia pastoris. Mol Biotechnol. 2000;16:23-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 629] [Cited by in RCA: 621] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 16. | Nakamura T, Nishizawa T, Hagiya M, Seki T, Shimonishi M, Sugimura A, Tashiro K, Shimizu S. Molecular cloning and expression of human hepatocyte growth factor. Nature. 1989;342:440-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1628] [Cited by in RCA: 1631] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 17. | Sambrook J, Russell DW. Molecular cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Third Edition, New York. 2001;. |
| 18. | Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8980] [Cited by in RCA: 9062] [Article Influence: 232.4] [Reference Citation Analysis (0)] |
| 19. | Sanchez JC, Wirth P, Jaccoud S, Appel RD, Sarto C, Wilkins MR, Hochstrasser DF. Simultaneous analysis of cyclin and oncogene expression using multiple monoclonal antibody immunoblots. Electrophoresis. 1997;18:638-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 20. | Jin H, Wyss JM, Yang R, Schwall R. The therapeutic potential of hepatocyte growth factor for myocardial infarction and heart failure. Curr Pharm Des. 2004;10:2525-2533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 42] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 21. | Yuan AH, Mei CL. effects of hepatocyte growth factor on synthesis of extracellular matrix and matrix metalloproteinases and tissue inhibitor of metalloproteinases in autosomal dominant polycystic kidney disease cyst-lining epithelial cells. Dier Junyi Daxue Xuebao. 2003;24:11-17. |
| 22. | Wu SG, Yu CL, Xu W, Guo YJ, and Ce XY. Expression and biological activity of human hepatocyte growth factor (HGF) by Chinese hamster ovary (CHO) cells. Diyi Junyi Daxue Xuebao. 1995;15:321-324. |
| 23. | Qiu YC, Zhu ZG, Xu JH, Xu W, Wu SG. Purification and identification of human recombinant hepatocyte growth factor expressed by CHO cells. Acad J First Mil Med Univ. 2001;21:332-333. |
| 24. | Wang MY, Yang YH, Lee HS, Lai SY. Production of functional hepatocyte growth factor (HGF) in insect cells infected with an HGF-recombinant baculovirus in a serum-free medium. Biotechnol Prog. 2000;16:146-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 25. | Bosch A, McCray PB, Walters KS, Bodner M, Jolly DJ, van Es HH, Nakamura T, Matsumoto K, Davidson BL. Effects of keratinocyte and hepatocyte growth factor in vivo: implications for retrovirus-mediated gene transfer to liver. Hum Gene Ther. 1998;9:1747-1754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 26. | Dang SY, Cheng NL, Niu B, Wang HZ, Zhao JB. Expression of human recombinant hepatocyte growth factor heavy chain in Escherichia coli. J Shanxi Med Univ. 2000;31:481-483. |
| 27. | Li XG, Gu YL, Zhu YS, Song HY. Molecular cloning of cDNA gene encoding human hepatocyte growth factor and its expression with Pichia. Pharmaceutical Biotechnology. 1997;4:193-197. |
Science Editor Wang XL and Guo SY Language Editor Elsevier HK