Published online Oct 21, 2005. doi: 10.3748/wjg.v11.i39.6152
Revised: April 8, 2005
Accepted: April 11, 2005
Published online: October 21, 2005
AIM: To identify the gene (s) related to the antagonistic activity of Enterobacter cloacae B8 and to elucidate its antagonistic mechanism.
METHODS: Transposon-mediated mutagenesis and tagging method and cassette PCR-based chromosomal walking method were adopted to isolate the mutant strain(s) of B8 that lost the antagonistic activity and to clone DNA fragments around Tn5 insertion site. Sequence compiling and open reading frame (ORF) finding were done with DNAStar program and homologous sequence and conserved domain searches were performed with BlastN or BlastP programs at www.ncbi.nlm.nih.gov. To verify the gene involved in the antagonistic activity, complementation of a full-length clone of the anrF gene to the mutant B8F strain was used.
RESULTS: A 3 321 bp contig around the Tn5 insertion site was obtained and an ORF of 2 634 bp in length designated as anrFgene encoding for a 877 aa polyketide synthase-like protein was identified. It had a homology of 83% at the nucleotide level and 79% ID/87% SIM at the protein level, to the admM gene of Pantoea agglomerans andrimid biosynthetic gene cluster (AY192157). The Tn5 was inserted at 2 420 bp of the gene corresponding to the COG3319 (the thioesterase domain of type I polyketide synthase) coding region on B8F. The antagonistic activity against Xanthomonas oryzae pv. oryzae was resumed with complementation of the full-length anrF gene to the mutant B8F.
CONCLUSION: The anrF gene obtained is related to the antagonistic activity of B8, and the antagonistic substances produced by B8 are andrimid and/or its analogs.
-
Citation: Yu XP, Zhu JL, Yao XP, He SC, Huang HN, Chen WL, Hu YH, Li DB. Identification of
anrF gene, a homology ofadmM of andrimid biosynthetic gene cluster related to the antagonistic activity ofEnterobacter cloacae B8. World J Gastroenterol 2005; 11(39): 6152-6158 - URL: https://www.wjgnet.com/1007-9327/full/v11/i39/6152.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i39.6152
Enterobacter cloacae strain B8 isolated from rice leaves was first recognized as an antagonistic bacterium of Xanthomonas oryzae pv. oryzae[1]. Its biological characteristics, antagonistic activity, and substances have been reported[2,3]. Satisfactory results have been obtained on anti-rice bacterial leaf brilliant in net house and in field[4]. Additionally, E. cloacae B8 is also antagonistic to many other plant bacterial pathogens such as Pseudomonas, Agrobacterium[3]. It has been reported that substances purified from E. cloacae B8 inhibit human (animal) bacterial pathogens such as MRSA[5,6].
Few but an E. cloacae strain EcCT-501R3 was reported to have antagonistic activity, which inhibits Pythium ultimumsporangium germination[7,8]. A competitive exclusion mechanism, competitive utilization of long-chain fatty acids of seed exudates, has been discovered and verified with transposon-mediated mutagenesis and tagging[9]. Previous researches on E. cloacae B8, however, showed that it produces antagonistic substances and differs from EcCT-501R3[1,3].
To elucidate the antagonistic mechanism of E. cloacae B8, transposon tagging[9-12] strategy was employed to clone DNA fragment related to its antagonistic activity. After a DNA fragment was cloned, cassette PCR-based chromosomal walking[13-15] was used to get the flanked unknown DNA. We have previously reported the cloning and analysis of an antagonistic-related DNA fragment[16,17] and the verification of loss of antagonistic activity of mutant strains, by Southern blotting analysis, as a result of the insertion of a single copy of Tn5[18]. Here, we report the cloning and identification of the full-length anrF gene and the verification of its participation in the antagonism of B8.
E. cloacae B8 (rifr) is the antagonistic bacterium isolated from rice leaves in our laboratory[3,19]. X. oryzae pv. oryzae strain Xcom3104 (rifr kanr) was used as an indicator bacterium of antagonistic activity. The suicide plasmid pZJ25 (cmr kanr mob::Tn5) and its host E.coli S17-1 (thi pro hsdR- hsdM+ recA RP4)[20] were gifts from Professor Jin-Sheng Wang of Nanjing Agricultural University, China. The cloning vector pBluescript SK+ (pBS) and its host E. coli DH5a were purchased from Stratagene. pFastBacTMHTA was purchased from Invitrogen. pMD18-T vector was a product of Takara. Restriction enzymes, T4 DNA ligase, CIAP, Taq DNA polymerase, dNTPs, DNA marker DL2000, marker 3 kb, PCR product purification kit, etc., were supplied by Promega, Takara, Sangon and/or Zeheng. Oligonucleotides and primers used in this research were synthesized in Bioasia Biotech.
The transfer of suicide plasmid pZJ25 from E. coli host S17-1 (donor) to E. cloacae B8 (receptor, rifr) was mediated with conjugation[21]. Tranposonated B8 clones (rifr kanr cms) were selected on plates with rifampicin and kanamycin. The antagonistic characteristics were then checked using Xcom 3104 (rifr kanr) as an indicator[19].
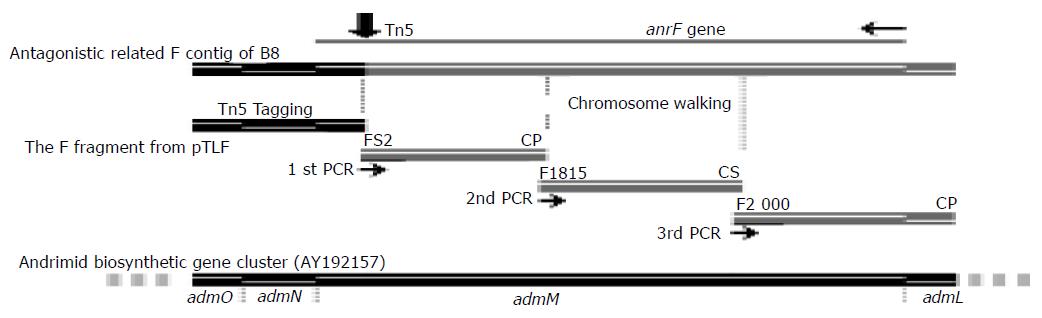
Genomic DNA of B8F, an antagonistic mutant strain of B8, was isolated, cut with BamHI and ligated to BamHI cut and dephosphorylated pBS. Recombinant clones were selected on plates with both kanamycin and ampicillin. Only one DNA fragment on the left of the insertion site was obtained with this step, as BamHI cut Tn5 into two fragments and the intact Kanrgene was in the 2.7 kb left fragment. The F fragment was a DNA fragment on the left of inserted Tn5 on B8F genome (Figure 1).
Two sets of cassettes and primers were adopted in our chromosomal walking experiments. The PstI cassette and its related CP primer were synthesized as previously described[12]. The Sau3AI cassette and its related CS primer and FS2, F1815 and F2000, the specific primers corresponding to known sequences, were newly designed and synthesized. They were as follows:
PstI cassette:
5'HO-AGATTCGGTGCGTGCTTGACTGCA-OH 3'
3'HO-TCTAAGCCACGCACGAACTG-OH 5'
Sau3AI cassette:
5'HO-CTGTGGTGGTTCCGATGTATG-OH 3'
3'HO-ACCACCAAGGCTACATACCTAG-OH 5'
Cassette primers:
CP primer: AGATTCGGTGCGTGCTTGAC
CS primer: CTGTGGTGGTTCCGATGTAT
Specific primers:
FS2: GTTTACTGGCCAGTTATTGTTG
F1815: CCACAGAACGTCTTGTCAT
F2000: CGGAATGAAGAGGGTAAGG
Genomic DNA was extracted from B8 and partially digested with Sau3AI or completely digested with PstI. DNA of 2-10 kb in length was recovered from gel and ligated to Sau3AI or PstI cassette to construct cassette genomic libraries. Related cassette primer (CS or CP) and specific primer were used to amplify target DNA of unknown region from the libraries. The PCR conditions were as follows: pre-denaturation for 5 min at 94 °C, and denaturation at 94 °C for 30 s, annealing at 52 °C for 60 s and extension at 72 °C for 5 min for 30 cycles, and a final extension at 72 °C for 10 min. Three rounds of PCR-based chromosomal walking, using PstI and Sau3AI cassettes and related primers by turns, were performed to cover the full length ofanrF gene (Figure 1).The full-length anrF gene was shown on the top (gray single line). The direction of the gene (narrow horizontal arrow, at right) and the Tn5 insertion site (thick vertical arrow) were marked. The long double line underneath stood for the F contig obtained in this research. The dark part on the left of Tn5 insertion site (the F fragment) was obtained with transposon tagging. The gray part on the right was obtained with chromosomal walking. The four separate lines below were four steps (one tagging and three rounds of PCR) to get the contig. Primers were marked on top of the three gray lines. The black double line at the bottom showed the corresponding genes of P. agglomerans andrimid biosynthetic gene cluster (AY192157).
The F fragment was sequenced, after subcloning of pTLF from M13 universal primer and P1 primer (CATGGAAG-TCAGATCCTGGA), a primer designed according to the sequence of the left edge of Tn5. PCR products of chromosomal walking were cloned into T-vector and sequenced using M13 universal primer. The DNA sequencing was done in Bioasia Biotech, Shanghai. DNA sequences obtained were edited, compiled, translated, and compared using DNAStar program. Homologous sequence and putative-conserved domain searches were done with BlastN and/or tBlastX and/or BlastP program atwww.ncbi.nlm.nih.gov.
Two primers were designed to amplify the full-length anrF gene. The upstream primer F21 (TTTTGTTGATGGAA-AGTCGCA) was located 144-126 bp up of the start codon. The downstream primer F2600 (CACCCAGCTGA-TGAAGTAAT) was located on opposite strand 73-56 bp down of the stop codon. The DNA fragments between the primers in the genomes of B8 and B8F were about 2 851 bp and 8.3 kb respectively. Genomic DNA of B8 or B8F was used as template. After 30 cycles of amplification, 6 mL of PCR products were applied to agarose gel electrophoresis to check the amplification. The amplified products were cloned and sequenced.
The full-length anrF gene amplified from B8 was cloned into pMD18-T vector and the resulting recombinant plasmid was designated as pMD-F. Gentamicin-resistance gene (Genr) was amplified with two primers (acc1-up: CGTGGAAACG-GATGAAG, acc1-down: ACCTGGCGGCGTTGTGACA) from pFastBacTMHTA and cloned into pMD18-T too. The resulting recombinant plasmid was designated as pMD-G. The Genr gene was recovered from pMD-G with HindIII and BamHI double cut and filled with klenow and inserted into XbaI cut and klenow filled pMD-F. The target plasmid with Genr and the full-length anrF gene was designated as pMD-FG.
Mutant E. cloacae B8F was cultured to an A600 about 0.6 in LB medium, and washed thrice in cold glycerol buffer. Plasmid pMD-FG was transformed into B8F with electroporation. The transformed bacteria were screened on plates with both ampicillin and gentamicin and verified by PCR amplification of anrF and Genr genes. The pMD-FG transformed strains were then inoculated on Wakimoto agar and cultured at 30 °C for 48 h, the resume of antagonistic characteristics was checked with the indication of X. oryzae pv. oryzae strain Xcom3104.
The suicide plasmid pZJ25 was transferred into E. cloacae B8 by conjugation with the help of donor E. coli S17-1. The conjugation and transposition ratio was about 5?0-6. A total of 1 500 transposed colonies (Rifr Kanr Cms) were obtained and two antagonistic mutant strains, one of which was designated as B8F, were selected.
A recombinant plasmid designated as pTLF, a plasmid with intact Kanr gene of Tn5 and a DNA fragment on the left of the insertion site (designated as F fragment), was isolated. The pTLF had two foreign BamHI fragments, one of which was about 3.5 kb in length and contained the target F fragment and a fragment of Tn5 (with the Kanr gene, about 2.7 kb in length). After subcloning, the 735 bp sequence of the F fragment was obtained with M13 universal sequencing primer and P1 primer.
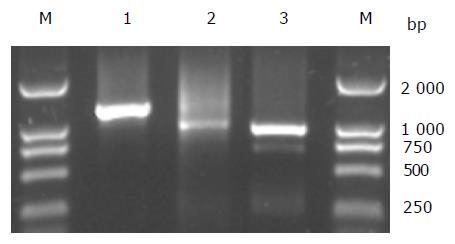
Only one DNA fragment on the left of insertion site was obtained with transposon tagging method. In order to know the full-length gene interrupted by Tn5 insertion, cassette PCR-based chromosomal walking method was adopted to clone DNA fragments on the right of insertion site. The framework of the cloning of the full-length anrF gene with transposon tagging and chromosomal walking was shown (Figure 1). With the use of specific primers (FS2, F1815, or F2000) and cassette primers (CP or CS) by turns, DNA fragments of unknown region were amplified from related cassette (PstI or Sau3AI) libraries. The PCR products from three different rounds of chromosomal walking were obtained (Figure 2). They were about 1.2 kb in PstI library (1st walking), about 950 bp in Sau3AI library (2nd walking), and about 900 bp in PstI library (3rd walking), respectively. The total sequence obtained with chromosomal walking was 2 586 bp.
A contig of 3 321 bp in length was obtained by transposon tagging and three rounds of chromosomal walking. Four ORFs, two complete and two partial were identified. One ORF, 2 634 bp in length, disrupted by the insertion of Tn5 in B8F was designated as anrF gene (accession no. AY633625) encoding for a 877 aa polyketide synthase-like protein. The Tn5 insertion site was at 2 420 bp of the anrF gene, and was at 214 bp before the stop codon.
BlastN search showed that the nucleotide sequence of anrF gene had a homology of 83% to that of admM gene of P. agglomerans andrimid biosynthetic gene cluster (AY192157). The alignment of anrF and admM genes is shown in Figure 3.
BlastP search showed that the deduced amino acid sequence encoded by the anrF gene was homologous to AdmM (79% ID/87% SIM) and many other proteins of polyketide/non-ribosomal peptide synthetase or polyketide synthase (type I) modules (about 28-38% ID/45-58% SIM). Conserved domain search showed that the N-terminal of the protein (about 1-410 aa) encoded for a polyketide synthase module (COG3321) or animal-type fatty-acid synthase (KOG1202) or ketoacyl synthase (pfam00109 and pfam02801) or 3-oxoacyl-(acyl-carrier-protein) synthase (FabB or COG0304, and KOG1394)-like domain. The middle sequence (about 550-610 aa) encoded for a phosphopantetheine attachment site (pfam00550)-like domain. The C terminal (about 640-877 aa) encoded for a thioesterase of type I polyketide synthase (COG3319, pfam00975, and COG3208)-like domain, which was disrupted by the insertion of Tn5 in B8F.
With F21 and F2600 as primers, no band was amplified in the reaction mixture using B8F genomic DNA as template, while a PCR product 2.8 kb in length, as expected, was observed in that of B8 (Figure 4). The 2.8 kb PCR product of B8 was further cloned and sequenced. The sequence result conformed the continuity of the anrF gene in B8.
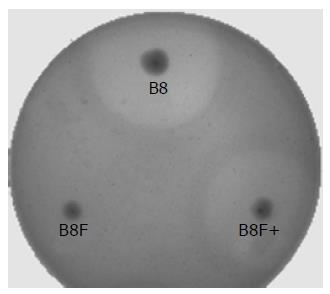
The pMD-FG, a plasmid with the full-length anrF and Genr genes, was constructed and transformed into the mutant E. cloacaeB8F. The antagonistic activity of pMD-FG transformed B8F against X. oryzae pv. oryzae was then resumed, though the antagonistic ring was a little bit small and ambiguous (Figure 5).
E. cloacae B8 is an antagonistic bacterium isolated from rice leaves, using X. oryzae pv. oryzae Xcom3104 as an indicator[1]. It has a strong antagonistic activity against X. oryzae pv. oryzae and satisfactory results have been obtained on anti-rice bacterial leaf brilliant in net house and in field[4]. Further researches showed that E. cloacae B8 and its antagonistic substances are also antagonistic to many other plant bacterial pathogens such as Pseudomonas, Agrobacterium[3], and human (animal) bacterial pathogens such as MRSA[5,6]. E. cloacae B8 and its antagonistic substances can be potentially used for the control of the rice bacterial leaf brilliant and many other bacterial diseases of plants, animals and humans.
It was reported that E. cloacae EcCT-501R3 has an antagonistic activity[7,8]. Two antagonistic-related genes for linoleic acid utilization, fadB and fadL, have been cloned using transposon-mediated mutagenesis and tagging. Further researches have verified that E. cloacae EcCT-501R3 inhibits Pythium ultimum sporangium germination by a competitive exclusion mechanism, the competitive utilization of long-chain fatty acids of seed exudates[9].
Our previous work, however, showed that B8 has an antagonistic mechanism different from what has been reported[1,3]. To elucidate the antagonistic mechanism and to clarify the antagonistic substances of E. cloacae B8, transposon tagging strategy[9-12] was employed to clone genes related to its antagonistic activity. With suicide plasmid pZJ25 carrying Tn5, two antagonistic mutant strains, one of which was named B8F, were selected. Tagging with Kanr gene of Tn5, the F fragment on the left of Tn5 insertion site was cloned in pTLF. Then, cassette PCR[13-15]-based chromosomal walking was applied to the amplification of unknown DNA fragments on the right of Tn5 insertion site. A contig of 3 321 bp in length was obtained after three rounds of chromosomal walking together with the F fragment from transposon tagging. One complete ORF of 2 634 bp in length, disrupted by the Tn5 insertion in B8F, was identified in the contig. It encoded for a 877 aa polyketide synthase-like protein and was designated as anrF gene. The Tn5 insertion site was at 2 420 bp of the anrF gene, and was at 214 bp before the stop codon, suggesting that the loss of antagonistic activity of mutant strain B8F was a result of the knock-out of anrF gene by Tn5.
To confirm this theory, the continuity of the anrF gene in the original B8 and its mutant B8F strains was checked. The complete anrF gene 2.8 kb in length was amplified from the B8 genome. The sequence result further confirmed the continuity of anrF gene in the original B8. The length of the DNA fragment between the primers in the genome of B8F was about 8.3 kb. However, no band was amplified from B8F. As the parameters set for amplification were suitable for amplification fragments 2.8 kb or less in length but not for fragments up to 8.3 kb.
In addition, plasmid pMD-FG with full-length anrF and Genr genes was constructed. After pMD-FG was transformed into B8F, a mutant strain of B8 knocked out by Tn5-mediated mutagenesis, the antagonistic characteristics were resumed, indicating that the anrF gene participated in the production of the antagonistic activity of B8.
BlastN search showed that the nucleotide sequence of the anrF gene had a homology of 83% to that of the admM gene ofP. agglomerans andrimid biosynthetic gene cluster (AY192157). BlastP search showed the deduced amino acid sequence encoded by the anrF gene was homologous to the AdmM (79% ID/87% SIM) and many other proteins of polyketide synthase/non-ribosomal peptide synthetase (about 28-38% ID/45-58% SIM). Domain search showed that the N-terminal of the protein encoded for a polyketide synthase module (COG3321) or animal-type fatty-acid synthase (KOG1202)-like domain. The middle sequence encoded for a phosphopantetheine attachment site (pfam00550)-like domain. And the C terminal encoded for a thioesterase of type I polyketide synthase (COG3319)-like domain, which was disrupted by the insertion of Tn5. It was this insertion that led to the loss of antagonistic activity of B8F, suggesting that the protein encoded by anrF gene is a polyketide synthase similar to AdmM of P. agglomerans, and that the antagonistic substances produced by B8 are andrimid and/or analogs of andrimid. This result is consistent with our early observations[1,3].
Andrimid and its analogs are antibiotic substances of the polyketide (pyrrolidinedione) family[22]. Only three different strains of bacteria, other than B8, have been reported to produce these substances so far. Frenenhagen et al[23] first reported the identification of andrimid, a new peptide antibiotic produced by an intracellular bacterial symbiont isolated from a brown planthopper. It has been reported that andrimid exhibits a moderate activity against Bacillus sp. and a very good activity against X. campestris, an early name of the pathogen of bacterial blight in rice plants[23]. Needham et al[24] reported that andrimid and Moiramides A-C are produced in culture by a marine isolate of Pseudomonas fluorescens. Singh et al[25] further showed that andrimid secreted from this strain exhibits antibacterial activity against both Gram-positive and Gram-negative bacteria. Oclarit et al[26] found that anti-Bacillus peptide antibiotic, an andrimid in the marine sponge, Hyatella species, is produced by an associated Vibrio species bacterium. Freiberg et al[22] reported that andrimid and its analogs act as selective inhibitors of bacterial acetyl-CoA carboxylase.
However, no genes related to the production of andrimid are published so far, except for the andrimid biosynthetic gene cluster of P. agglomerans (AY192157). The complete genome of AY192157 is 28 485 bp in length and encodes for 21 genes, from admA, admB to admU. The anrF gene cloned in this research has a homology of 83% to admM. Furthermore, the arrangement and the sequences of other ORFs or non-coding regions around the anrF gene have shown that the F contig obtained in this research is an equivalent of a part of AY192157 (data not shown). P. agglomerans (formerly Erwinia herbicola) is a closely related but nonpathogenic bacterium of E. amylovora[27]. It usually accompanies E. amylovora in the wild and produces a family of antibiotics[28-30] to inhibit fire blight, a devastating disease of rosaceous plants such as apple and pear caused by the latter bacterium. E. cloacae B8 has been isolated from rice leaves as an antagonistic bacterium of X. oryzae pv. oryzae. However, both E. cloacae and P. agglomerans are members of the Enterobacteriacea family. Whether this is the reason why the anrF and admM genes have a high similarity, or whether the equivalents of andrimid biosynthetic genes are similar or identical in bacteria other than Enterobacteriacea family, including Pseudomonas and Vibrio remains to be studied.
| 1. | Chen WL, Ge WX, Li DB. A study on Enterobacter cloacae B8, Bacillus subtillus B826 and their antagonistic substance to Xanthomonas campestris pv. oryzae. Zhejiang Nongye Daxue Xuebao. 1990;16:61-67. |
| 2. | Xu YP, Zang RC, Chen WL, Lou YC. Promoting plant growth and IAA analysis of Enterobacter cloacae B8 fermentation liquid. Zhejiang Daxue Xuebao. 2001;27:282-284. |
| 3. | Chen WL. Antagonistic substances produced by bacterial strains to Xanthomonas oryzae pv. oryzae. Ph. D. thesis. Zhejiang Unviersity. 2001;59-66. |
| 4. | Chen WL, Xu P, Gong FH, Li DB. Study on coloninzation of Enterobacter cloacae B8x on rice leaves and its control of rice bacterial leaf blight (Xanthomonas oryzae pv. oryzae). Nongye Shengwujishu Xuebao. 1994;2:61-66. |
| 5. | Zhang R, Chen WL, Zhu BR. The antagonistic activities of the extract of Enterobacter clocae B8 to methicillin-resistant Staphylococcus aureus. Zhonghua Weishengwuxue He Mianyixue Zazhi. 2003;23:172. |
| 6. | Ya-Ping J, Wei-liang C, Rong Z, Bai-rong Z. The biological activity of antibacterial substance produced by Enterobacter cloacae B8. Rinsho Biseibutshu Jinsoku Shindan Kenkyukai Shi. 2003;13:115-120. [PubMed] |
| 7. | van Dijk K, Nelson EB. Inactivation of seed exudates stimulants of Pythium ultimum sporangium germination bybiocontrol strain of Enterobacter cloacae and other seed-associated bacteria. Soil Biol Biochem. 1998;29:351-355. |
| 8. | Kageyama K, Nelson EB. Differential inactivation of seed exudate stimulation of Pythium ultimum sporangium germination by Enterobacter cloacae influences biological control efficacy on different plant species. Appl Environ Microbiol. 2003;69:1114-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | van Dijk K, Nelson EB. Fatty acid competition as a mechanism by which Enterobacter cloacae suppresses Pythium ultimum sporangium germination and damping-off. Appl Environ Microbiol. 2000;66:5340-5347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 37] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 10. | Wei JF, Zhang SG, Ma YC. Transposon Tn5-induced INA- Mutants in INA Bacteria. Yunan Nongye Daxue Xuebao. 2002;17:1-4. |
| 11. | Mills D. Transposon mutagenisis and its potential for studying virulence genes in plant pathogens. Ann Rev Phytopathol. 1985;23:297-320. [DOI] [Full Text] |
| 12. | Kumar A, des Etages SA, Coelho PS, Roeder GS, Snyder M. High-throughput methods for the large-scale analysis of gene function by transposon tagging. Methods Enzymol. 2000;328:550-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 13. | Kilstrup M, Kristiansen KN. Rapid genome walking: a simplified oligo-cassette mediated polymerase chain reaction using a single genome-specific primer. Nucleic Acids Res. 2000;28:E55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 14. | Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith JC, Markham AF. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990;18:2887-2890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 457] [Cited by in RCA: 501] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 15. | Zhang Z, Gurr SJ. Walking into the unknown: a 'step down' PCR-based technique leading to the direct sequence analysis of flanking genomic DNA. Gene. 2000;253:145-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 46] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Yu XP, Zhu JL, Yao XP, He SC, Huang HN, Chen WL, Li DB. Cloning and analysis of the antagonistic related genes of Enterobacter cloacae B8. Chin Sci Bull. 2004;49:1370-1375. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 17. | Yu XP, Zhu JL, Yao XP, He SC, Huang HN, Chen WL, Li DB. Cloning and analysis of the antagonistic related genes of Enterobacter cloacae B8. Chin Sci Bull. 2004;49:1370-1375. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 18. | Huang HN, Yu XP, Gong HF, Xu P, Chen WL, Li DB. Cloning and detecting of the antagonistic related genes of Enterobacter cloacae B8 with transposon tagging method. Zhejiang Daxue Xuebao 2005, in press. . |
| 19. | Xu P, Chen WL, Zhu WG, Li DB. A drug resisitant mutant of antagonistic bacterium Eterobacter cloacae B8. Zhejiang Nongye Daxue Xuebao. 1992;18:115-119. |
| 20. | Li P, Long JY, Huang YC, Zhang Y, Wang JS. The avrXa3, a new member of avrBs3 family isolated from Xanthomonas oryzae pv. oryzae is an avirulent gene of double function. Ziran Kexue Jinzhan. 2004;14:767-781. |
| 21. | Wang JS, He CY, Ye HY. Tn5-induced virulence gene mutants and their biochemical properties of Xanthomonas campestris pv. oryzae. Zhiwu Bingli Xuebao. 1992;22:217-223. |
| 22. | Freiberg C, Brunner NA, Schiffer G, Lampe T, Pohlmann J, Brands M, Raabe M, Häbich D, Ziegelbauer K. Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors with antibacterial activity. J Biol Chem. 2004;279:26066-26073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 157] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 23. | Frenenhagen A, Tamura SY, Kenny PTM, Komura H, Naya Y, Nakanishi K. Andrimid, a new peptide antibiotic produced by an intracellular bacterial symbiont isolated from a brown planthopper. J Am Chem Soc. 1987;109:4409-4411. [RCA] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 124] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 24. | Needham J, Kelly MT, Ishige M, Andersen RJ. Andrimid and moiramides A-C, metabolites produced in culture by a marine isolate of the bacterium Pseudomonas fluorescens: structure elucidation and biosynthesis. J Org Chem. 1994;59:2058-2063. [RCA] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 81] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Singh MP, Mroczenski-Wildey MJ, Steinberg DA, Andersen RJ, Maiese WM, Greenstein M. Biological activity and mechanistic studies of andrimid. J Antibiot (Tokyo). 1997;50:270-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 26. | Oclarit JM, Okada H, Ohta S, Kaminura K, Yamaoka Y, Iizuka T, Miyashiro S, Ikegami S. Anti-bacillus substance in the marine sponge, Hyatella species, produced by an associated Vibrio species bacterium. Microbios. 1994;78:7-16. [PubMed] |
| 27. | Gavini F, Mergaert J, Beji A, Mielcarek C, Izard D, Kersters K, de Ley J. Transfer of Enterobacter agglomerans (Beijerinck 1988) Ewing and Fife 1972 to Pantoea gen. nov. as Pantoea agglomerans comb. nov. and description of Pantoea dispersa sp. nov. Int J Syst Bacteriol. 1989;39:337-345. [RCA] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 253] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 28. | Wright SA, Zumoff CH, Schneider L, Beer SV. Pantoea agglomerans strain EH318 produces two antibiotics that inhibit Erwinia amylovora in vitro. Appl Environ Microbiol. 2001;67:284-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 101] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 29. | Giddens SR, Houliston GJ, Mahanty HK. The influence of antibiotic production and pre-emptive colonization on the population dynamics of Pantoea agglomerans (Erwinia herbicola) Eh1087 and Erwinia amylovora in planta. Environ Microbiol. 2003;5:1016-1021. [RCA] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 30. | Wodzinski RS, Paulin JP. Frequency and diversity of antibiotic production by putative Erwinia herbicola strains. J Appl Bacteriol. 1994;76:603-607. [RCA] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.5] [Reference Citation Analysis (1)] |
Science Editor Guo SY Language Editor Elsevier HK
Co-first-authors: Xu-Ping Yu and Jun-Li Zhu|
Co-correspondents: Xu-Ping Yu and De-Bao Li