INTRODUCTION
A candidate tumor suppressor gene, inhibitor of growth 1 (ING1), has recently been cloned and mapped on human chromosome 13q33-34[1-3]. It has at least three exons and two introns. Its three alternative spliced transcripts encode p47ING1a, p33ING1b, and p24ING1c protein, respectively[4]. p33ING1b is a nuclear protein, which physically interacts with p53, and cooperates with it in many ways[2,5]. Overexpression of p33ING1b could efficiently block the growth of cells and induce apoptosis[6-8]. Conversely, inhibition of its expression by antisense constructs could promote cells growth and protect cells from apoptosis[7]. Downregulation of p33ING1b has been reported to be closely related with occurrence of various cancers, such as head and neck[4], esophageal[9], lung[10], gastric[11,12], bladder[13], ovary[14], breast[14,15], liver cell cancer[16,17], and so on. Genetic alterations, such as mutation or allelic imbalance, have been found in some cancers listed above. To the author's knowledge, only a few studies of ING1 gene on colorectal cancer have been reported as yet, and the results are different from each other[18]. Moreover, there is some lack of knowledge about p47ING1a expression in colorectal cancer till now. Accordingly, in order to evaluate the effect and significance of ING1 gene in carcinogenesis and progression of human sporadic colorectal cancer, we examined mRNA expression, mutation, and LOH of ING1 gene by using RT-PCR, PCR-SSCP, and PCR-RFLP.
MATERIALS AND METHODS
Patients and tissues
The tumor and matched normal mucosal tissues (at least 10 cm away from tumor margin) were obtained from 35 patients with sporadic colorectal cancer, who underwent surgical resection in our hospital, from December 2002 to March 2003. The patients consisted of 20 males and 15 females with the mean age of 59 years (range, 25-79 years). All the diagnoses were verified by pathological diagnosis. Sixteen were colon and 19 were rectal carcinomas with Dukes' stages as follows: stage A (2 cases); stage B (15 cases); stage C (13 cases); and stage D (5 cases). All the specimens were frozen in liquid nitrogen after removal, followed by storage at -80 °C.
RNA and DNA extraction
Total RNA and genomic DNA were isolated from the specimens using TRIzol reagent (Invitrogen, USA) and spin column genomic DNA isolation kit (BBI, Canada) according to the manufacturer's instructions. The integrity of total RNA and genomic DNA were checked on 10 g/L agarose gel.
RT-PCR
ING1 mRNA expression in tumor and matched normal tissues was examined using RT-PCR. RNase-free DNase I was used to remove DNA contamination at first. Then, cDNA was synthesized using first strand cDNA synthesis kit, following the manufacturer's instructions. At last, PCR was performed in 20 μL reaction mixtures comprising 2 μL of cDNA, 2.5 μL of 10×buffer, 1.5 μL of 2.5 mol/L deoxynucleotide triphosphate mixture (dNTP mixture), 2 U of Taq DNA polymerase (TaKaRa, Dalian, China), 0.5 μL of 10 mol/L each primer of ING1, 0.5 μL of 10 mol/L primer of GAPDH, and 12 μLof sterilized ddH2O. Intron-spanning primers were: for p47ING1a, 5'-CGCATCTTT-GCTGACCCGA-3'and 5'-GCCTTCTTCTCCTTG-GGTGT-3'(444 bp); and for p33ING1b, 5'-CTCCATCGAG-TCCCTGCCTT-3'and 5'-GCCTTCTTCTCCTTGG-GTGT-3'(480 bp). Primers for GAPDH were 5'-CGG-AGTCAACGGATTTGGTGCGTAT-3'and 5?AGCCTT-CTCCATGGTGGYGAAGAC-3'(306 bp). The samples were amplified through 32 cycles, each amplification consisting of denaturation at 94 °C for 1 min, primer annealing at 62 °C for 1 min and extension at 72 °C for 1 min. Cycles were preceded by incubation at 94 °C for 5 min to ensure full denaturation of the target gene, followed by an extra incubation at 72 °C for 10 min to ensure full extension of the products. Primers for GAPDH were added to PCR tubes at the end of the 10th cycle. Products of RT-PCR were electrophoresed on 20 g/L agarose gel. The light intensities of DNA bands were scanned by electrophoretic image analysis system (Bio-Rad Gel DOC 2000, USA). Reproducibility was confirmed by processing all samples at least twice.
PCR-SSCP
PCR reactions were carried out in a volume of 20 μL containing 100 ng of genomic DNA, 2.5 μL of 10×buffer, 1.5 μLof 2.5 mol/L dNTP mixture, 2 U of Taq DNA polymerase (TaKaRa, Dalian, China), 0.5 μL of 10 mol/L each primer, and 14 μL of sterilized ddH2O. The samples were amplified through 32 cycles, each amplification consisting of denaturation at 94 °C for 30 s, primer annealing at 62 °C or 64 °C for 30 s and extension at 72 °C for 1 min. Finally, the samples underwent an extra incubation at 72 °C for 10 min. The sequences of ING1 primers used for PCR-SSCP: exon 1, 5'-TGCAGTGCTATTTTT-TGAGGGG-3'and 5'-CGCCCCCGCCCATCCATCA-3' (248 bp); exon 2a, 5'-ACGCCTGTCCTTCTTGCCCC-3'and 5'-CTTGCCGCTGTTGCCCGCTG-3' (272 bp); exon 2b, 5'-TTCGAGGCGCAGCAGGAGCT-3'and 5'-CTTGGCCTTCTTCTCCTTGGG-3'(210 bp); exon 2c, 5'-TGAGCCCCACGCACGAGAAG-3'and 5'-CAGCA-ACCACGACCACGACG-3'(242 bp); exon 2d, 5'-CCTCC-CCATCGACCCCAACG-3'and 5'-ACATTTTACACT-CCTTGCACCTCA-3'(327 bp). Ten microliters of PCR products was mixed with 10 μL of loading dye (950 mL/L formamide, 20 mol/L EDTA, 0.5 g/L bromophenol blue, and 0.5 g/L xylene cyanol), heat denatured at 98 °C for 10 min, chilled on ice, put onto 120 g/L polyacrylamide gel, and run at a constant rate of 12 V/cm for 10-14 h at variable temperatures. The DNA bands were detected by silver staining as previously described[19]. Reproducibility was confirmed by processing all samples at least twice.
LOH analysis
We examined genomic DNA for LOH by PCR-SSCP using five microsatellite markers (D13S158, D13S278, D13S285, D13S779, and D13S796), which are located in the ING1 locus. The primer pairs were available through the internet genome database (http://www.gdb.org). PCR was performed in 20 μL of reaction mixture comprising 100 ng of genomic DNA, 2.5 μL of 10×buffer, 1.5 μL of 2.5 mol/L dNTP mixture, 2 U of Taq DNA polymerase (TaKaRa, Dalian, China), 0.5 μL of 10 mol/L each primer, and 14 μL of sterilized ddH2O. The samples were amplified through 32 cycles, each amplification consisting of denaturation at 62 °C for 30 s, primer annealing at 52 °C or 54 °C for 30 s and extension at 72 °C for 1 min. Finally, the samples underwent an extra incubation at 72 °C for 5 min. Ten microliters of PCR products was mixed with 10 μL of loading dye as described above, heat denatured for 10 min at 98 °C, chilled on ice, put onto 120 g/L polyacrylamide gel containing 8 mol/L urea, and run at a constant rate of 25 V/cm for 3-4 h at room temperature. The DNA bands were visualized by silver staining. Reproducibility was confirmed by processing all samples at least twice. LOH was scored, if the heterozygous alleles showed at least 50% reduced intensity in tumor DNA bands as compared with the corresponding normal ones.
Statistical analysis
The SPSS 10.0 software package was used for all statistical analyses. Differences in expression of ING1 mRNA between the different groups were compared by using independent t-test. A P value less than 0.05 was considered statistically significant.
RESULTS
NG1 mRNA expression detected by RT-PCR
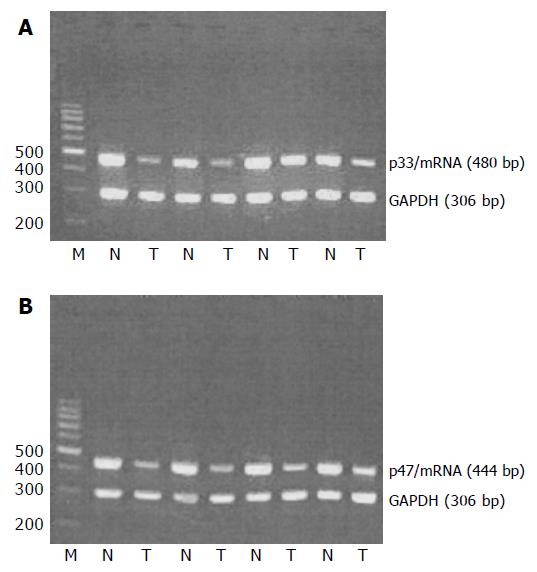
All the samples of tumor tissues and normal tissues showed ING1 mRNA expression. Moreover, the average ratios of light intensities of p33ING1b mRNA and p47ING1a mRNA expression in the cancerous tissues were significantly lower than those in normal tissues (0.52 vs 1.28, P<0.01; 0.51 vs 1.21, P<0.01, respectively; Figure 1); difference between the two mRNA splices was not significant in the matched tissues.
Figure 1 RT-PCR analysis of p33/ING1 and p47/ING1 mRNA expressions.
N: normal tissue; T: tumor tissue (A and B).
The average ratios of light intensities of p33ING1b mRNA and p47ING1a mRNA expression in the cancerous tissues of Dukes'stages C and D were significantly lower than those in cancerous tissues of Dukes'stages A and B (0.38 vs 0.65, P<0.01; 0.40 vs 0.63, P<0.01, respectively).
Detection of ING1 gene mutation
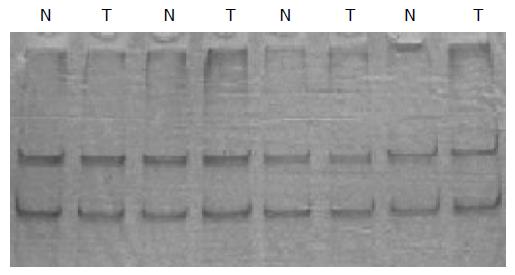
To investigate whether the ING1 gene is the target of functional loss in tumors, we searched for mutation in the encoding regions of the gene in all 35 samples of colorectal cancer by using PCR-SSCP. However, no mutation was found (Figure 2).
Figure 2 No mutation of ING1 gene was detected by PCR-SSCP (fragment exon 2a).
LOH analysis
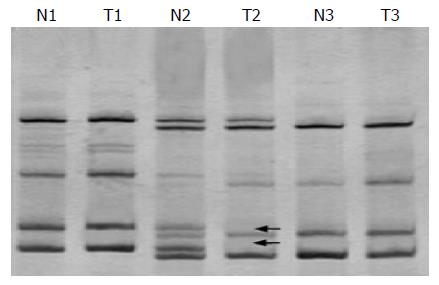
To investigate whether allelic imbalance of terminal regions of chromosome 13q is reserved, we examined DNA from 35 pairs of matched tumor tissues and normal tissues for losses at five microsatellite markers (D13S158, D13S278, D13S285, D13S779, and D13S796). But only four cases of LOH (11.4%) were observed, two in D13S158, one in D13S278 and one in D13S285 (Figure 3).
Figure 3 Electrophoretic picture of LOH analysis (LOH is seen in T2).
DISCUSSION
In this study, we found downregulation of p33ING1 mRNA, and p47ING1 mRNA expressions in the cancerous tissues of sporadic colorectal cancer, and the later Dukes'stage they were in, the less mRNA expression they presented. Our results are consistent with the previous data on head and neck[4], esophageal[9], lung[10], gastric[11,12], breast[14,15], and liver cell[16,17]cancers, neuroblastomas[20], and astrocytoma[21], but not with the other studies on colorectal cancer[18]. The precise mechanisms of tumor suppression by p33ING1 and p47ING1 remain unclear. Previous several reports thought that p33ING1 might inhibit cells growth and promote apoptosis through both p53-dependent[5,8,17,21] and -independent pathways[21-24]. One is: p33ING1 could stabilize p53 via disrupting the regulation of p53 by MDM2[23], and promote acetylation of p53 through inhibition of hSIR2[21,24]. Another one is: p33ING1 and p47ING1 could directly repress transcription promoters, and upregulate p21WAF1 promoter activity[21]. Moreover, p33ING1 could cooperate with p53. We, therefore, think that reduced expressions of p33ING1 and p47ING1 may lead to decreased inhibition of cell cycle and apoptosis, and strongly associated with the carcinogenesis and progression of human sporadic colorectal cancer.
Some previous studies have reported about several mis-sense mutations and silent changes in esophageal[9], head and neck[4], esophagogastric junction[25], gastric[11], skin[26], pancreatic[27], and breast[14] cancers. These mutations were all located in PDH finger and/or nuclear localization signal in the COOH-terminal half of ING1, and might lead to loss of the functions of ING1. But the rate of mutations was rare or no mis-sense mutation was found in oral squamous cell carcinoma[28], lymphoid malignancies[29], colorectal carcinoma[18], and hematological malignancies[30]. In our study, we could not detect mis-sense mutation of ING1 gene. Although PCR-SSCP analysis was reported to be less than 100% sensitive for mutation detection, this result suggests that mutation of ING1 occur with a very low frequency in sporadic colorectal cancer. We, hereby, propose that gene mutation is not the main reason for the downregulation of ING1 expression.
Loss of heterozygosity (LOH), i.e., loss of one allele at a constitutional heterozygous locus, indicates the probability of loss of a tumor suppressor gene, which might promote neoplastic progression and metastasis. LOH is a critical event in the development of human cancers. It is thought to have resulted from either a large deletion or recombination between homologous alleles during repair of DNA double-strand breaks. These types of genetic alterations may produce mutations in some gene mutation assay. Previous studies demonstrated that high rates of LOH were found in human chromosome 13q33-34 region[4,9,31], such as the rates in esophageal/head and neck squamous cell cancers were 58.9% (20/34) and 68% (23/34), respectively[4,9]. Because the gene encoding ING1 is located in 13q33-34 region, we detected for LOH in this region using five microsatellite markers (D13S158, D13S278, D13S285, D13S779, and D13S796), which are at different locations on chromosome 13q33-34. In our study, the rate of LOH was only 11.4% (4/35), which is obviously lower than those in the previous reports. LOH at ING1 locus and the vicinity may affect the function of ING1, but these might not be the main reason for the downregulation of ING1 expression as well.
In conclusion, downregulation of p33ING1 and p47ING1 mRNA expressions in human sporadic colorectal cancer is substantial, relating with the carcinogenesis and progression, but mutation and LOH of gene might not be the main reason for the downregulation of ING1 expression. The reason for loss of function of ING1 gene is very complex. We speculate that the alterations at transcriptional or post-transcriptional levels, such as acetylation, methylation, ubiquitination, or phosphorylation, might be involved in the downregulation of ING1 gene. Further studies are needed to elucidate the possibility.