Published online Sep 7, 2005. doi: 10.3748/wjg.v11.i33.5169
Revised: February 15, 2005
Accepted: February 18, 2005
Published online: September 7, 2005
AIM: p53-Inducible ribonucleotide reductase small subunit 2 (p53R2) encodes a 351-amino-acid peptide, which catalyzes conversion of ribonucleoside diphosphates to the corresponding deoxyribonucleotides required for DNA replication and repair. A recent study reported that a point mutation (G/T) in the p53 binding sequence in a colon cancer cell line completely impaired p53R2 protein activity.
METHODS: We screened the p53R2 gene coding regions and a regulatory region which contains a p53 binding sequence in 100 patients with colorectal adenoma and 100 control subjects using PCR, cold SSCP, and direct DNA sequencing.
RESULTS: Although we did not identify genetic variation in all nine exons, four regulatory-region variants were found, of which three were single nucleotide polymorphisms (SNPs) (nt 1 789 C/G, nt 1 928 A/G, 1 933 T/C), and one was 20 bp insertion which replaced a ATTTT between nt 1 831 and 1 835. Additionally, we determined the frequency of these p53R2 variants in a recently concluded case-control study of incident sporadic colorectal adenomas (163 cases and 210 controls).
CONCLUSION: Although more detailed functional characterizations of these polymorphisms remain to be undertaken, these polymorphic sites may be useful for identifying alleles associated with mis-splicing, additional transcript factors and, more generally, in cancer-susceptibility association studies.
- Citation: Deng ZL, Xie DW, Bostick RM, Miao XJ, Gong YL, Zhang JH, Wargovich MJ. Novel genetic variations of the p53R2 gene in patients with colorectal adenoma and controls. World J Gastroenterol 2005; 11(33): 5169-5173
- URL: https://www.wjgnet.com/1007-9327/full/v11/i33/5169.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i33.5169
The p53 gene is a major target for genetic alterations or biochemical activations in human cancer. However, the mechanism by which p53 exerts its effects or alters other signaling systems is largely unknown. In recent years, a number of specific p53 target genes have been discovered that are likely to be involved in downstream target effects of altered p53 protein, including cell-cycle arrest, apoptosis, and tumor suppression. Recently, an important p53 downstream target gene, p53-inducible ribonucleotide reductase small subunit 2 (p53R2) consisting of nine exons and a p53-banding sequence in intron 1, was identified in cancer cell lines[1]. Human ribonucleotide reductase catalyzes the rate-limiting step in the production of deoxyribonucleotide triphosphate (dNTP), which is required for DNA replication and repair[2]. This enzyme is also regulated by the cell cycle[3]. The R2 subunit is made late in the G1 phase before DNA replication, and disappears in late S or early G2. p53R2 is a gene that is essential for DNA repair, and loss of function of this gene results in dysfunctional repair mechanisms[1]. Polymorphisms in either the regulatory or coding regions of the p53R2 gene may affect its expression and protein function, altering an individual’s response to DNA damage, and thus affecting the risk of colorectal cancer. We screened the p53R2 gene coding regions and a regulatory region, which contains a p53 binding sequence, in 100 patients with colorectal adenoma and 100 control subjects for single-nucleotide polymorphisms and other mutations using PCR, cold SSCP, and direct DNA sequencing. Herein, we report several novel genetic variations designated as DdeI, NdeI, HhaI (according to restriction enzymes cut sites), and 20 bp insertion. Subsequently, we determined the allele frequencies of the observed polymorphisms in 163 cases of incident sporadic colorectal adenoma and 216 colonoscopy negative controls.
From 1994 to 1997, the Markers for Adenomatous Polyps (MAP) case-control study was conducted to assess the validity of colonic epithelial cell proliferation as a biomarker of risk for incident sporadic colorectal adenomatous polyps. Prior to the beginning of the study, MAP was approved by the Institutional Review Board of Wake Forest University School of Medicine in accordance with an assurance filed with and approved by the Department of Health and Human Services. Informed consent was obtained from each participant.
Eligibility criteria for study subjects consisted of English speaking adults from 30 to 74 years of age, of either sex, and of all races who were scheduled for elective outpatient colonoscopy by four large gastroenterology practices in Winston-Salem and Charlotte, North Carolina. Patients increased in number over a 24-mo period. Cases were identified as eligible colonoscopy patients who were determined to have a study index of pathologist-confirmed incident adenomatous polyps according to the criteria adapted from the National Polyp Study[4]. Controls consisted of all eligible colonoscopy patients with no previous history of adenomatous polyps and who were found to be free of adenomatous polyps. Persons with familial polyposis, Gardner’s syndrome, ulcerative colitis, Crohn’s disease, bowel resection, newly diagnosed recurrent adenomatous polyps, and incident colon cancer were excluded, as they had a history of cancer other than non-melanoma skin cancer. Blood was drawn and stored at -70 °C for possible later measurement of various genotypes.
Among all three clinical sites, 2 246 colonoscopy patients were identified. Of these, 669 were eligible on initial screening (eligibility rate 29.8%), and of these 633 were willing to discuss the study, 617 of these were contacted, and 417 of these signed consent and had study colonoscopies (consent rate 63.1%). Of the 417 participants, 259 had some types of polyp, and of these, 179 had adenomatous polyps. Nine of the 417 total patients were subsequently determined ineligible for the study and 8 additional patients had incident colon cancer and were not eligible for the primary case-control analyses; thus, 400 patients were available for genotypic analysis. Of these 400 patients, viable DNA was isolated from 373 (163 cases and 210 controls) for genotyping.
Genomic DNA was obtained from stored white blood cells (WBCs) digested in 500 μL of lysis buffer (50 mmol/L Tris-HCl, pH 8.5, 1 mmol/L EDTA, 0.2% SDS, 200 g/mL proteinase K) overnight at 55 °C with shaking. The digestion was precipitated directly with isopropanol and the pellets were washed with 70% ethanol. The genomic DNA pellets (50-100 μg) were dissolved in 300-800 μL of TE buffer, of which about 1 μL was used for each PCR reaction.
The primers (Table 1) and PCR product sizes were designed following published genomic DNA sequences (GenBank accession no.: NC_000008). We designed primers that were 18-25 bases long with a 40-60% GC content and an annealing temperature (Tm) around 52-68 °C. To obtain the optimal conditions for PCR amplification, we ran a set of PCRs with various concentrations of Mg++, template DNA, dNTPs, and primers for each fragment.
| Number | Name | Sequence | PCR product (bp) |
| 1 | Exon 1 (F) | 5’-GGAGAGTCACTCAATGGAC-3’ | 302 |
| 2 | Exon 1 (R) | 5’-CCTGCAACTTGCAATCTAAC-3’ | |
| 3 | Exon 2 (F) | 5’-GGAACACTGCACTATAGGATG -3’ | 291 |
| 4 | Exon 2 (R) | 5’-CCCTGACTTCCTTAGATG-3’ | |
| 5 | Exon 3 (F) | 5’-GCTAAAGGAGAACAAAAG-3’ | 218 |
| 6 | Exon 3 (R) | 5’-CCAAACACTTTGAGAAGAAC-3’ | |
| 7 | Exon 4 (F) | 5’-GCTCAGAAACTTCCATACTTG-3’ | 336 |
| 8 | Exon 4 (R) | 5’-CGATAATGCTGATGTCCAG-3’ | |
| 9 | Exon 5 (F) | 5’-CGTACTGGATAGTCACAC -3’ | 287 |
| 10 | Exon 5 (R) | 5’-CGAGATCGTGCCACTGCAC-3’ | |
| 11 | Exon 6 (F) | 5’-GGCACTAACTTGTGTATTTTG-3’ | 284 |
| 12 | Exon 6 (R) | 5’-CTAGAAAAACATTCCATTCC-3’ | |
| 13 | Exon 7 (F) | 5’-GGTACAAACATCAGAGAAAG-3’ | 224 |
| 14 | Exon 7 (R) | 5’-CCATCACCATATGCAAATAG-3’ | |
| 15 | Exon 8 (F) | 5’-GGTCCTTGCTTTCCTATA-3’ | 239 |
| 16 | Exon 8 (R) | 5’-CGAAAGTCCTCTTTCTA-3’ | |
| 17 | Exon 9 (F) | 5’-GCAGAGGAAAATAGACTAC-3’ | 317 |
| 18 | Exon 9 (R) | 5’-GGTTTTGAGAAACCTGAC-3’ | |
| 19 | Intron 1 (F) | 5’-CAGGACTCAGTAGAGGAGCT-3’ | 550 |
| 20 | Intron 1 (R) | 5’-CACAGGGTCAGTGACACTG-3’ |
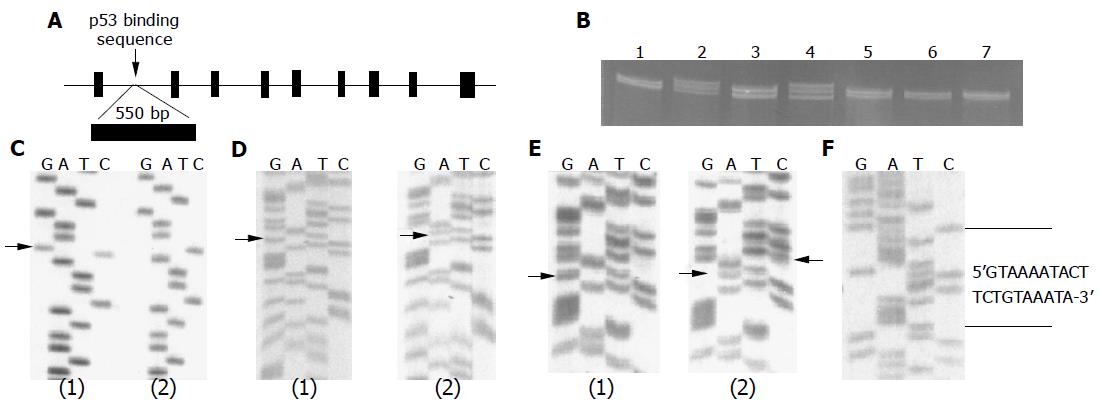
Five microliters of the PCR product was mixed with 15 μL denatured buffer [0.4 μL 1 mol/L MMH (methylmercury hydroxide), 12.6 μL 1.25×TBE, 2.0 μL 15% Ficoll containing 0.1% bromophenol blue, and 0.1% cyanol xylene blue], and then incubated for 10 min at room temperature, heated for 5 min at 95 °C, and put on ice before being loaded on 20% TBE polyacrylamide gel and electrophoresed in a Novex Thermoflow system for 4-5 h at 10-15 °C at 300 V. The gel was stained in 1×TBE with 5 μg/mL ethidium bromide for 10 min and visualized on a UV lamp (Figure 1B). The shift bands were cut and re-amplified for direct sequencing.
For direct sequencing, the PCR products amplified from genomic DNA were purified and directly used for sequencing. DNA was sequenced with the Thermal Sequenase Cycle Sequencing Kit from Amersham Life Science (Cleveland, OH, USA) according to manufacturer’s instructions. In cycle sequencing, the thermal cycle consisted of 2 min at 94 °C, 45 cycles of 1 min at 94 °C, 1 min at 60 °C, and 1 min at 72 °C. The reaction mixture was loaded on a 6% polyacrylamide sequencing gel made from Sequagel DNA Sequencing Solutions (National Diagnostics, GA, USA).
Since polymorphisms at the nt 1 789 G and nt 1 923 C alleles created DdeI and HhaI restriction enzyme cut sites, respectively, each PCR product was subjected to DdeI and HhaI restriction enzymes digestion prior to electrophoresis for determining genotype frequencies. The DNA fragments were then separated using 3% 2:1 Nusiev/SeaKern agarose gel. The allele types were determined for DdeI analysis as follows: three fragments of 6, 171, and 373 bp for the C allele, four fragments of 6, 97, 171, and 276 bp for the G allele; and five fragments of 6, 171, 276, and 373 bp for the C/G allele. The allele types were determined for HhaI analysis as follows: three fragments of 550 bp for the T allele, two fragments of 299 and 251 bp for the C allele; and three fragments of 550, 299, and 251 bp for the T/C allele nt 1 928 A/G, the polymorphism does not have a suitable restriction enzyme site. Therefore, we changed an A nucleotide to a T in the near 3 end of the forward primer. This change combines with the wild type allele (A) to create an NdeI cutting site, but does not do so with the variant allele (G). The PCR products (10 μL) were subjected to NdeI restriction enzyme. Bands for the wild-type (A/A) allele were cut into 246 and 26 bp; the G/G variant allele showed at 272 bp; and the heterozygote (A/G) allele was cut into 272, 246, and 26 bp fragments (Figure 2).
All statistical inquiries were conducted using R language version 1.9.0 from http://www.R-project.org. Homogeneity test was used to check the non-dependency between genotypes and the frequencies of the p53R2 gene variants between the patients with colorectal adenoma and the control groups. Consistency of the genotype frequencies with the Hardy-Weinberg equilibrium was also tested by χ2-analysis.
We screened nine exons and a regulatory region in first intron of p53R2 gene in 200 subjects including 100 patients with colorectal adenoma and 100 control subjects for mutation identification. No mutation was found in any of the nine exons; however, three SNP variations (nt 1 789 C/G, nt 1 928 A/G, and nt 1 933 T/C) were identified in the first intron of the p53R2 gene by cold SSCP (Figure 1B), and confirmed by direct DNA sequencing (Figures 1C-E and Table 2). A 20 bp variant, which is a short insertion variation that replaced 5 bp between nt 1 831 and nt 1 835 (Figure 1F and Table 2), was also found (Table 2). These polymorphisms are summarized in Figure 2.
| Cases (n = 163) | Controls (n = 210) | P | |||
| n | % | n | % | ||
| DdeI | |||||
| CC | 143 | 87.7 | 185 | 85.6 | |
| GG | 1 | 0.6 | 2 | 0.9 | |
| CG | 19 | 11.7 | 29 | 13.4 | 0.82 |
| NdeI | |||||
| GG | 131 | 80.4 | 178 | 82.4 | |
| AA | 1 | 0.6 | 6 | 2.8 | |
| AG | 30 | 18.4 | 31 | 14.4 | 0.25 |
| HhaI | |||||
| TT | 159 | 97.6 | 210 | 97.2 | |
| CT | 4 | 2.4 | 6 | 2.8 | |
| CC | 0 | 0 | 0 | 0 | 0.9 |
| Insertion | |||||
| (+) | 4 | 2.4 | 0 | 0 | - |
Restriction enzyme mapping showed two restriction enzymes that could be used for population screening for nt 1 789 C/G (DdeI) and nt 1 933 T/C (HhaI) variations. We also designed a modified RFLP (NdeI) method for population screening for the nt 1 928 A/G alteration (Figure 2). Therefore, C/G, T/C, and A/G polymorphisms were designated DdeI, HhaI, and NdeI, respectively. Then, frequencies of these variations were determined in all of the 373 subjects (cases = 163, control = 210) from case-control study (Table 2). Since each of these four variations is close to the p53 binding sequence, they may play important roles in maintaining a three-dimensional structure with the p53 binding sequence. Of the four variations, the HhaI polymorphism occurred at a very low frequency (2.78%). The 20 bp variant was found in only 4 patients, all adenoma cases. The distribution of DdeI genotypes C/C, C/G, G/G, and G/G combined with C/G were 87.7%, 11.7%, 0.6% and 12.3% in cases, compared to 85.7%, 13.4%, 0.9% and 14.3% in controls, respectively. The distribution of NdeI genotypes G/G, G/A, A/A, and A/A combined with G/A were 80.0%, 18.4%, 1.23% and 19.6% in cases, compared with 82.8%, 14.4%, 2.79%, 17.2% in controls, respectively. All polymorphisms were in Hardy-Weinberg equilibrium in all populations, with the exception of the 20 bp variant, which was found in 4 patients only, all with colorectal adenoma. There is no statistical difference between the cases and control subjects (Table 2).
p53-Inducible ribonucleotide reductase small subunit 2 (p53R2) is an important p53 downstream target gene that is essential for DNA repair[1]. It has been demonstrated that p53R2 has an important role in delivering dNTP and maintaining the dNTP pool in response to DNA damage. Loss of function of this gene results in dysfunctional repair mechanisms.
Single-strand conformation polymorphism (SSCP) is a technique for detecting known and unknown genetic variation using PCR products[5]. Using this method about 70-90% of unknown mutations can be detected. Single strands assume a specific folded conformation, when double-stranded PCR product is denatured to single strand. These conformers are separated by non-denaturing polyacrylamide gel electr-ophoresis. A single base mutation can cause a change in the folded conformation compared to the wild type and can change the separation pattern. This means that a change in separation pattern compared to that of the wild type indicates a mutation.
Using these methods, we here report mutation screening of the p53R2 gene, identification of four novel genetic variations in a regulatory region, their allele frequencies in both normal and colorectal adenoma populations, and the predicted functional impact of resulting transcriptional changes. Since each of these four mutations is close to the p53 binding sequence, these variations may play an important role in maintaining a three-dimensional structure with the p53 binding sequence. As of May 2004, only two polymorphisms of the p53R2 gene (-88 C/A and 8 bp insertion) have been reported[6,7]. We searched the available literature and the NCBI SNP database, and found no published information on these novel variants. To our knowledge, this is the first report of allele frequencies of novel p53R2 polymorphisms in a population consisting of patients with colorectal adenoma and controls.
Somatic mutation screening of p53R2 has been previously investigated in cancer. On the whole, two studies investigated the genetic variation of p53R2 in gastric and urothelial carcinoma[8,9]. Byun et al, did not detect any somatic variations in the p53R2 gene in 105 gastric carcinomas, including 15 cell lines[8]. One non-synonymous coding variant (Glu136/Asp) was identified in urothelial carcinoma; however, the frequency was rare (1/108)[9]. It was concluded that p53R2 is not a critical target of genetic inactivation in tumorigenesis.
Genetic polymorphisms have been identified in many genes related to DNA synthesis and repair, such as thymidylate synthase (TS)[10,11], MTHFR[11,12], and XRCC1[13,14]. Genetic polymorphisms could affect gene expression, enzyme function, or the protein’s interaction with environmental and nutritional factors to modify the risk of neoplasia as we have demonstrated for other polymorphisms[15]. An 8 bp tandem repeat and a -88 C/A polymorphism were discovered in the 5’-untranslated region of the p53R2 gene[8,9]. Three polymorphisms in 3’-untranslated region of p53R2 gene were also identified[16]. Since we focused only on exons and the first intron regulatory regions, we did not investigate these polymorphisms.
As physical locations of these polymorphisms are so close to the p53 binding sequence as described above, it will be quite interesting to investigate their associations with cancer risk. Because polymorphic alleles at loci as close together as these are likely to be transmitted together, it will also be of interest to define haplotypes for the p53R2 gene.
In summary, we have reported newly identified polym-orphisms in the p53R2 gene, and shown allele frequencies of the new variants in a case-control study population consisting of 163 patients with colorectal adenoma and 210 control subjects. Since p53R2 is important in response to DNA damage, and thus may also be in colon cancer prevention, genetic polymorphisms that may alter the expression of this gene are of interest. Further studies of the functional impact of these polymorphisms are needed. It would also be of great interest to investigate their possible association with colorectal cancer risk.
| 1. | Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 654] [Cited by in RCA: 672] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 2. | Jordan A, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 1998;67:71-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 588] [Cited by in RCA: 568] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 3. | Björklund S, Skog S, Tribukait B, Thelander L. S-phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry. 1990;29:5452-5458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 142] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 4. | O'Brien MJ, Winawer SJ, Zauber AG, Gottlieb LS, Sternberg SS, Diaz B, Dickersin GR, Ewing S, Geller S, Kasimian D. The National Polyp Study. Patient and polyp characteristics associated with high-grade dysplasia in colorectal adenomas. Gastroenterology. 1990;98:371-379. [PubMed] |
| 5. | Hongyo T, Buzard GS, Calvert RJ, Weghorst CM. 'Cold SSCP': a simple, rapid and non-radioactive method for optimized single-strand conformation polymorphism analyses. Nucleic Acids Res. 1993;21:3637-3642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 231] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 6. | Smeds J, Kumar R, Hemminki K. Polymorphic insertion of additional repeat within an area of direct 8 bp tandem repeats in the 5'-untranslated region of the p53R2 gene and cancer risk. Mutagenesis. 2001;16:547-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 7. | Smeds J, Nava M, Kumar R, Hemminki K. A novel polymorphism (-88 C& gt; A) in the 5' UTR of the p53R2 gene. Hum Mutat. 2001;17:82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 8. | Byun DS, Chae KS, Ryu BK, Lee MG, Chi SG. Expression and mutation analyses of P53R2, a newly identified p53 target for DNA repair in human gastric carcinoma. Int J Cancer. 2002;98:718-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Hayashi H, Furihata M, Kuwahara M, Kagawa S, Shuin T, Ohtsuki Y. Infrequent alteration in the p53R2 gene in human transitional cell carcinoma of the urinary tract. Pathobiology. 2004;71:103-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 10. | Horie N, Aiba H, Oguro K, Hojo H, Takeishi K. Functional analysis and DNA polymorphism of the tandemly repeated sequences in the 5'-terminal regulatory region of the human gene for thymidylate synthase. Cell Struct Funct. 1995;20:191-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 396] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 11. | Chen J, Giovannucci E, Kelsey K, Rimm EB, Stampfer MJ, Colditz GA, Spiegelman D, Willett WC, Hunter DJ. A methylenetetrahydrofolate reductase polymorphism and the risk of colorectal cancer. Cancer Res. 1996;56:4862-4864. [PubMed] |
| 12. | Ma J, Stampfer MJ, Giovannucci E, Artigas C, Hunter DJ, Fuchs C, Willett WC, Selhub J, Hennekens CH, Rozen R. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 1997;57:1098-1102. [PubMed] |
| 13. | Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604-608. [PubMed] |
| 14. | Ratnasinghe DL, Yao SX, Forman M, Qiao YL, Andersen MR, Giffen CA, Erozan Y, Tockman MS, Taylor PR. Gene-environment interactions between the codon 194 polymorphism of XRCC1 and antioxidants influence lung cancer risk. Anticancer Res. 2003;23:627-632. [PubMed] |
| 15. | Lewis RC, Bostick RM, Xie D, Deng Z, Wargovich MJ, Fina MF, Roufail WM, Geisinger KR. Polymorphism of the cyclin D1 gene, CCND1, and risk for incident sporadic colorectal adenomas. Cancer Res. 2003;63:8549-8553. [PubMed] |
| 16. | Ye Z, Parry JM. The discovery and confirmation of single nucleotide polymorphisms in the human p53R2 gene by EST database analysis. Mutagenesis. 2002;17:361-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
Co-first-authors: Zong-Lin Deng and Da-Wen Xie
Science Editor Guo SY Language Editor Elsevier HK