Published online Jul 28, 2005. doi: 10.3748/wjg.v11.i28.4337
Revised: December 1, 2004
Accepted: December 3, 2004
Published online: July 28, 2005
AIM: To investigate the effect of NS398 on the metastasis-associated gene expression in LoVo colorectal cancer cells.
METHODS: LoVo cells were treated with NS398 at the concentration of 100 mmol/L for 24 and 48 h respectively. Total RNA was extracted with TRIzol reagents and reverse transcribed with Superscript II and hybridized with cDNA microarray (containing oncogenes, tumor suppressor genes, signal transduction molecules, adhesive molecules, growth factors, and ESTs) fabricated in our laboratory. After normalization, the ratio of gene expression of NS398 treated to untreated LoVo cells was either 2-fold up or 0.5-fold down was defined as the differentially expressed genes. Semi-quantitative RT-PCR was used to validate the microarray results.
RESULTS: Among the 447 metastasis-associated genes, 9 genes were upregulated and 8 genes were downregulated in LoVo cells treated with NS398 for 24 h compared to untreated cells. While 31 genes were upregulated and 14 genes were downregulated in LoVo cells treated with NS398 for 48 h. IGFBP-5, PAI-2, JUN, REL, BRCA1, and BRCA2 might be the new targets of NS398 in treatment of colorectal cancer.
CONCLUSION: NS398 might exert its anti-metastasis effect on colorectal cancer by affecting several metastasis-associated gene expression.
- Citation: Gao XQ, Han JX, Huang HY, Song B, Zhu B, Song CZ. Effect of NS398 on metastasis-associated gene expression in a human colon cancer cell line. World J Gastroenterol 2005; 11(28): 4337-4343
- URL: https://www.wjgnet.com/1007-9327/full/v11/i28/4337.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i28.4337
N-[2-(cyclohexyloxy)-4-nitrophenyl] methanesulfonamide (NS398) is a highly selective cyclooxygenase-2 (COX-2) inhibitor. Its mechanism in anticancer effects involves many signal pathways. One is induction of apoptosis of different tumor cells. NS398 inhibits the viability of colon cancer cell lines by apoptosis by the release of cytochrome C from mitochondria and by the activation of caspase-9 and caspase-3 and cleavage of poly (ADP-ribose) polymerase. Cytochrome C pathway plays an important role in NS398-induced apoptosis in colon cancer cell lines[1]. NS398 may also suppress the growth of tumor cells by inhibiting the cell cycle progression. It can increase the inhibitor of cell cycles p27Kip1 by inhibiting protein degradation to suppress the proliferation of human lung cancer cells, and this in turn is caused by modulating p27Kip1 proteolysis. Non-steroid anti-inflammatory drugs (NSAIDs) suppress the expression of chymotrypsin-like catalytic subunits (LMP5, LMP7, and LMP2), but do not directly block enzymatic activity and inhibit proteasome activity. Reverse transcriptase-competitive PCR and promoter activity assays showed that this inhibition occurred at the transcriptional level[2]. NS398 can exert its anti-angiogenesis and anti-metastasis effects by inhibiting the expression of vascular endothelial growth factor (VEGF)[3]. Its inhibitory effects on the metastasis in vitro and in vivo are mainly mediated by regulating the matrix metalloproteinase (MMP) family components. NS398 can inhibit the invasiveness of prostate cancer by reducing the release of MMP-2 and MMP-9 and increase of TIMP-2 but not TIMP-1[4]. NS398 inhibits MMP-2 mRNA expression and also decreases the amount of MMP-2 in human lung cancer cells. Additionally, this COX-2 inhibitor attenuates the degrading activity of MMP-2. The synthesis and processing of MMP-2 was significantly suppressed by NS398. NS398 directly inhibits MMP-2 promoter activity. However, the inhibitory effect of NS398 is not fully dependent on inhibition of COX-2 because a high concentration of NS398 is needed to suppress MMP-2 expression and addition of prostaglandin E2 only partially reverses the action of NS398[5].
NS398 and aspirin also upregulate RECK mRNA level in CL-1 human lung cancer cells. Additionally, NSAIDs increase RECK protein level which was associated with reduction of MMP-2 activity. NSAID-activated RECK expression may not be mediated via inhibition of COXs because addition of prostaglandin E2 (PGE2) cannot counteract the effect of NSAIDs and overexpression of COX-2 cannot downregulate RECK[6]. To promote the application of NS398 in the treatment and chemoprevention of colorectal cancer, and observe whether it has other target genes, we detected the effect of NS398 on the expression of metastasis-associated genes in colorectal cancer cell lines by cDNA microarray.
A total of 447 cDNA clones were obtained from Research Genetics (Invitrogen, Life Technologies, USA). E.coli with inserted metastasis-associated genes were cultured with Luria-Bertain culture medium supplemented with ampicillin (50 mg/L in final concentration) or chloromycin (170 mg/L) in InnovaTM4330 refrigerated incubator shaker (New Brunswick Scientific, USA) at the speed of 250 r/min overnight at 37°C. Clone plasmids were extracted with Edge BioSystems (Gaithersburg, MD, Germany). Clone inserts were PCR-amplified from the plasmids with M13 vector-specific universal primer (M13F: 5'-GGT GTA AAA CGA CGG CCA GTG-3'; M13R: 5'CAC ACA GGA AAC AGC TAT G-3') in 96-well PCR microtiters. The PCR products were purified with protocols published[7], and resuspended in Arrayit spot solution. The purified PCR products were printed on silanated slides (CEL Associates, Houston, TX, USA) with Cartisan PixSys 5500 robot (Cartesian Technologies, Irvine, CA, USA) and cDNA microarrays were UV-cross-linked at 3 500 mJ using Cl-1000 ultraviolet cross-linker (Stratagene). Microarrays were post-processed according to protocol online[8].
LoVo cells were grown in the culture incubator at 37°C with 50 mL/L CO2 in RPMI 1640 (Life Technologies, USA) supplemented with 10% neonatal bovine serum. After the cells were cultured to 60-70% confluence, 12 mL of NS398 dissolved in dimethyl sulfoxide (Me2SO) was added to make the final concentration 100 mmol/L and further cultured for 24 and 48 h respectively. The same amount of Me2SO was added to the control.
LoVo cells treated with NS398 or Me2SO were lysed with TRIzol (Life Technologies Inc., Rockville, MD, USA) according to the manufacturer's protocol and total RNA was extracted and stored at -80°C. The concentration of total RNA were measured with a biophotometer (Eppendorf AG22331, Hamberg, Germany) and the 260/280 ratio of RNA was 1.8-2.0.
Probes were prepared as described previously[9,10] with some modifications. First strand cDNA was synthesized by priming 10mg total RNA with 6 mg random hexamers (Life Technologies Inc., Rockville, MD, USA) by heating at 70°C for 10 min, snap-cooling on ice for 30 s and placed at room temperature for additional 5-10 min. Reverse transcription was performed in the presence of 500 mmol/L each of dATP, dCTP and dGTP, 200 mmol/L aminoallyl-dUTP (Sigma Chemical Co., St. Louis, MO, USA), 300 mmol/L dTTP, 1 first strand buffer, 10 mmol/L dithiolthreitol, and 400 U superscript II (Life Technologies) in 30 mL reaction at 42°C overnight. Reactions were quenched with 0.5 mol/L EDTA and RNA template was hydrolyzed by addition of 10mL NaOH of 1 mol/L followed by heating at 70°C for 10 min. Reactions were neutralized with 10 mL 1 mol/L HCl and cDNA was purified with Amicon Microcon YM100 (Millipore Corporation, Bedford, MA, USA) according to the manufacturer's protocol. cDNA was dried in speed vacuum concentrator 5301 (Eppendorf, Germany) and resuspended in 4.5 mL 0.1 mol/L (pH 9.0) sodium carbonate buffer. Aliquot of Cy3 NHS ester dye (Amersham Pharmacia Biotech, UK) was dissolved in 4.5 mL Me2SO (1mg dye from one tube was dissolved in 73 mL of Me2SO and aliquot in 16 tubes, dried in speed vacuum and stored at 4°C) and added to the resuspended cDNA and reactions were incubated at room temperature in the dark for 1 h. Coupling reactions were quenched by addition of 41 mL 0.1 mol/L sodium acetate (pH 5.2), and unincorporated dye was removed using QIAquick PCR purification kit (Qiagen, Germany) following manufacturer's instructions.
Each slide was printed with duplicate microarrays. Slides were pre-hybridized in 1% BSA, 5 SSC, 0.1% SDS for 45 min, washed twice in de-ionized double distilled H2O and 2-propanol and air-dried and used in 1 h. Fluorescent cDNA probes were dried in speed vacuum and resuspended in 10 mL hybridization buffer p5 mL formamide, 2.5 mL 20 SSC, 1.0 mL reagent grade double distilled water (RGDD H2O), 0.5 mL 2% SDS and 1 mL human cot-1DNA]. Probes were denatured at 100°C water bath for 2 min and cooled at room temperature for 5 min. Room temperature probes of NS398 treated and untreated group were applied to the duplicate microarrays on the same pre-hybridized microarrays, covered with hybridized coverslip (Sigma) and placed in the hybridization chamber (Corning). Hybridizations was carried out at 42°C water bath for 20-22 h followed by washing in 2 SSC and 0.1% SDS for 3 min, 1 SSC for 2 min and 0.2 SSC for 1 min and 0.05 SSC for 10 s, and dried by spin at horizontal plate centrifuge at 90 r/min for 4 min. Microarrays were scanned using a ScanArray 4000 (Packard Bioscience, PE, USA) dual color confocal laser scanner. Data were saved as paired TIFF images.
Spots were identified and local background subtracted in the QuantArray 3.0. In the first step, a grid consisting of square cells was drawn around each array element. Spot segmentation was then performed using a fixed segmentation method that uses the distribution of pixel intensity to separate probable signal from background and a binary threshold approach to identify spots, followed by a procedure to exclude disconnected features. Raw intensity for each element was obtained by first excluding saturated pixels, then summing all remaining pixel intensities inside the spot contours. The area outside the spot contour but inside the cell was used to calculate local background. Background per pixel was estimated as a median of the pixels in this area and multiplied by the spot area to give an estimated spot background value. In the final step, this integrated background value was subtracted from the raw integrated spot intensity to produce the background-subtracted integrated intensities used for further analysis. Furthermore, a quality control filter was used to remove questionable array features. Two criteria for spot rejection were the spot shape deviating greatly from a circle and a low signal to noise ratio. Spots for which the ratio of area to circumference deviated by more than 20% from the value for an ideal circle and spots containing less than 50% of pixels above the median background values were flagged and eliminated from further consideration. The spot intensity above blank plus 2SD was used for the final analysis. Then the data were normalized to total with software supplied by the manufacturer. The two fold up- or down-regulated genes were shown in red or green respectively.
The upregulated gene IGFBP-5 was measured by RT-PCR to verify the microarray results. RT-PCR was performed on MJ-PTC200 DNA engine using TaKaRa two-step reaction with protocols supplied by the manufacturer. The primers for IGFBP-5 were forward: 5'-TTG CCT CAA CGA AAA GAG C-3', reverse: 5'-AGA ATC CTT TGC GGT CAC A3'[11]. The primers for b-actin were forward: 5'- AAG TAC TCC GTG TGG ATC GG -3', reverse: 5'- TCA AGT TGG GGG ACA AAA AG -3'[12]. PCR was performed at 94°C for 2 min, and 30 cycles at 94°C for 30 s, at 50°C for 30 s and at 72°C for 60 s and a final extension at 72°C for 5 min. PCR products were electrophoresed on 1% agarose gel. Images were captured with Alpha ImageTM 2000. The band density was measured with the software supplied by the same system.
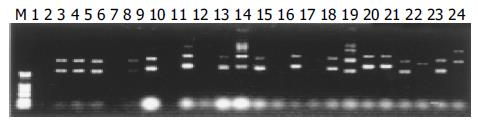
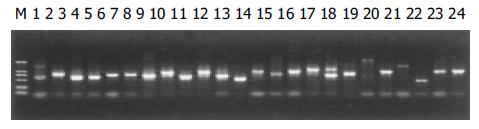
The plasmids were extracted by Edge biosystems plasmids extraction kit (Gaithersberg, MD, Germany). The extracted plasmids were run on 1% agarose gel. The results shown in Figure 1 are representative of the 447 clones. PCR amplification of the 447 clones for the inserts is shown in Figure 2. The single band amplification rate was 93%.
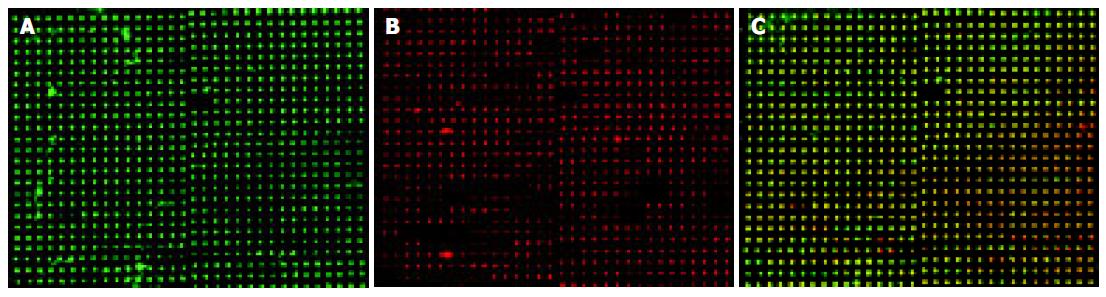
To identify metastasis-associated genes affected by NS398, LoVo cells were treated with 100 mmol/L NS398 for 24 and 48 h respectively. The representative image is shown in Figure 3. Image A represents the hybridized results of untreated cells. Image B represents the hybridization results of the NS398-treated LoVo cells and image C is the overlay image of NS398-treated to control LoVo cells.
After treatment with NS398 for 24 h, 9 genes were upregulated and 8 genes downregulated (Table 1). The downregulated genes included lipocalin 2 (oncogene 24p3) (LCN2), intercellular adhesion molecule 5, telencephalin (ICAM5), v-jun avian sarcoma virus 17 oncogene homolog (JUN), v-rel avian reticuloendotheliosis viral oncogene homolog (REL). Five genes after being treated for 24 h were still highly expressed until 48 h and one gene was still inhibited until 48 h.
| Accession number | Gene descriptions | Ratio |
| Not found | Hs.23723, “ESTs” (346390) | 2.67±1.66 |
| Not found | Hs.106513, “EST” (348242) | 2.31±0.67 |
| BC040844 | Hs.198253, NS1-associated protein 1 (435598) | 1.96±0.25 |
| BC011714 | Hs.9605, heterogeneous nuclear ribonucleoprotein D-like (190165) | 2.08±0.46 |
| Not found | Hs.277401, ESTs (35363) | 2.04±0.13 |
| NT_024524 | Hs.277704, PIBF1 gene product (124966) | 2.29±0.81 |
| BC007674 | Hs.180414, CD24 antigen (small cell lung carcinoma cluster 4 antigen) (115306) | 1.98±0.22 |
| NM_003127 | Hs.87497, spectrin, alpha, non-erythrocytic 1 (alpha-fodrin, 3021698) | 2.27±0.53 |
| Not found | Hs.272073, ESTs (1296662) | 1.95±0.24 |
| BT007404 | Hs.8037, CD24 antigen (small cell lung carcinoma cluster 4 antigen, 124098) | 0.49±0.04 |
| K01500 | Hs.18443, alpha-1-antichymotrypsin (117439) | 0.51±0.05 |
| Not found | Hs.45209, EST(2118886) | 0.49±0.04 |
| NM_005564 | LCN2 (oncogene 24p3, 595821) | 0.47±0.17 |
| NM_001022 | Hs.43913, ribosomal protein S19 (453963) | 0.53±0.06 |
| NM_003259 | Intercellular adhesion molecule 5, telencephalin (ICAM5, 180864) | 0.36±0.14 |
| NM_002228 | v-jun avian sarcoma virus 17 oncogene homolog (JUN, 823612) | 0.51±0.07 |
| NM_002908 | v-rel avian reticuloendotheliosis viral oncogene homolog (REL, 2723459) | 0.45±0.06 |
After 48 h of treatment more genes were regulated by NS398. Of the 447 genes analyzed, 31 genes were upregulated and 14 genes downregulated (Table 2). IGFBP-5, APAF, LAMP2, CLCA2, laminin gamma3, HOXA1, WNT2, N-cadherin, PAI-2, TIMP could inhibit the metastasis of tumor.
| Accession number | Gene description | Ratio |
| Not found | Hs.23723, “ESTs” (346390) | 3.41±1.76 |
| Not found | Hs.106513, “EST” (348242) | 3.05±1.88 |
| BC040844 | Hs.198253, NS1-associated protein 1 (435598) | 2.51±0.53 |
| AF257505 | Hs.23317, butyrophilin, subfamily 3, member A2 (219410) | 2.12±0.56 |
| NT_033899 | Hs.28043, KIAA0712 gene product (219914) | 2.13±0.40 |
| BC011714 | Hs.9605, heterogeneous nuclear ribonucleoprotein D-like (190165) | 2.34±0.51 |
| M62782 | Hs.22907, human insulin-like growth factor binding protein 5 (IGFBP5) mRNA (31397) | 2.15±0.43 |
| NM_003127 | Hs.87497, spectrin, alpha, non-erythrocytic 1 (alpha-fodrin, 3021698) | 2.14±0.13 |
| BC008005 | Hs.116459, nucleotide binding protein 2 (E coli MinD like, 2498589) | 2.01±0.29 |
| Not found | Hs.43913, ESTs (2498857) | 2.20±0.28 |
| Not found | Hs.272073, ESTs (1296662) | 2.33±0.43 |
| BM508995 | Hs.106513, ESTs, highly similar to proteasome (H sapiens, 1302647) | 2.47±0.76 |
| NM_000624 | Hs.150580, alpha-1-antichymotrypsin (1322220) | 2.14±0.35 |
| AF248734 | Hs.87497, apoptotic protease activating factor (963055) | 2.28±0.45 |
| BC002965 | Lysosomal-associated membrane protein 2 (LAMP2), transcript variant LAMP2A (134418) | 2.34±0.69 |
| NM_006536 | Chloride channel, calcium activated, family member 2 (CLCA2, 781187) | 2.39±0.42 |
| M90657 | Tumor-associated antigen L6 (1964132) | 2.85±1.58 |
| Not found | Spliceosome associated protein 145 (1964680) | 2.28±0.34 |
| AW674474 | Putative insulin-like growth factor ii associated (229316) | 2.32±0.50 |
| AF041835 | Laminin, gamma 3 (LAMC3, 2497685) | 2.16±0.48 |
| AB019987 | Chromosome-associated polypeptide-c (2597847) | 2.06±0.09 |
| AW277011 | Putative vacuolar protein sorting-associated protein c (2744695) | 2.20±0.64 |
| BC032547 | Homeo box A1 (HOXA1, 3923611) | 2.27±1.18 |
| NM_003391 | Wingless-type MMTV integration site family member 2 (WNT2, 149373) | 2.27±0.28 |
| BC027948 | c-fos induced growth factor (VEGF D, FIGF, 160946) | 2.15±0.27 |
| NM_001792 | Cadherin 2, N-cadherin (neuronal, CDH2, 3617894) | 2.03±0.10 |
| NM_001964 | Early growth response 1 (EGR1, 182411) | 2.61±0.47 |
| AF071400 | Plasminogen activator inhibitor, type II (arginine-serpin, PAI2, 323255) | 2.41±0.39 |
| NM_002447 | Macrophage stimulating 1 receptor (c-met-related tyrosine kinase, MST1R, 586698) | 2.45±0.62 |
| NM_003254 | Tissue inhibitor of metalloproteinase 1 (erythroid potentiating activity, | |
| collagenase inhibitor, TIMP1, 771755) | 2.02±0.18 | |
| NM_003182 | Tachykinin, precursor 1 (substance K, substance P, neurokinin 1, neurokinin 2, neuromedin L, | 2.22±0.30 |
| neurokinin alpha, neuropeptide K, neuropeptide gamma, TAC1), transcript variant beta (784179) | ||
| BC004986 | Hs.77202, ribosomal protein S25 (178052) | 0.49±0.06 |
| BC035128 | Hs.23317, Max-interacting protein (130696) | 0.45±0.05 |
| Not found | Hs.23954, ESTs (132543) | 0.51±0.06 |
| AJ001810 | Hs.106513, pre-mRNA cleavage factor Im (25 ku) | 0.48±0.01 |
| BC053521 | 66834, Hs.76847, spectrin, alpha, non-erythrocytic 1 (alpha-fodrin, 31230) | 0.47±0.15 |
| M64716 | Hs.234726, ribosomal protein S25 (4932742) | 0.46±0.05 |
| BC032589 | No, beta-2-microglobulin (1907327) | 0.48±0.11 |
| Not found | Hs.45209, EST (2118886) | 0.47±0.11 |
| NT_024524 | Hs.15058, PIBF1 gene product (1596167) | 0.50±0.05 |
| NM_001779 | CD58 antigen (lymphocyte function-associated antigen 3, CD58, 490368) | 0.50±0.15 |
| NM_002228 | v-jun avian sarcoma virus 17 oncogene homolog (JUN, 823612) | 0.41±0.11 |
| NM_020979 | Adaptor protein with pleckstrin homology and src homology 2 domains (APS, 3056093) | 0.49±0.17 |
| NM_000059 | Breast cancer 2, early onset (BRCA2, 3850805) | 0.30±0.10 |
| NM_001223 | Caspase 1, apoptosis-related cysteine protease (interleukin 1, beta, convertase, CASP1, 3858119) | 0.48±0.09 |
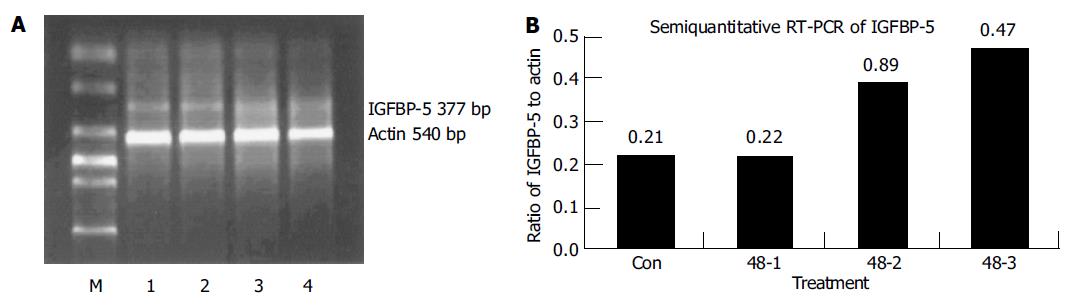
IGFBP-5 gene expression was validated with TAKARA version 2.1 RT-PCR kit. The expression level was measured with alpha InnoTechTM 2000 spot density method. The NS398-treated LoVo cells expressed more IGFBP-5 mRNA (Figure 4). The relative expression ratio to b-actin was 0.21, 0.22, 0.39, and 0.47 for the control and the three NS398-treated replicate experiments. The IGFBP-5 mRNA level in NS398-treated LoVo cells to untreated LoVo cells was 1.05, 1.85, and 2.23 respectively for the three replicate experiments. It was in accord with the microarray results.
NS398 is a highly selective COX-2 inhibitor. Its anticancer effects have been linked to cell apoptosis and inhibition of MMP and anti-angiogenesis. NS398 also exerts its synergistic effect with radiotherapy for the treatment of head and neck squamous cell cancer (HNSCC) by inhibiting radiation-induced expression of COX-2[12] and increases the sensitivity of chemotherapy by enhancing the expression of cyclin-dependent kinase inhibitors p21 (Waf1) and p27 (Kip1), promoting apoptosis of tumor cells, therefore makes the cells stay in G1 arrest[13].
cDNA microarray has been widely used in the screening of drug targets. To identify whether NS398 had other target genes in the treatment of colorectal cancer, we treated LoVo cells with NS398 and the expression of metastasis-associated genes were measured with microarray. The results showed that NS398 influenced the expression of some metastasis genes. After being treated for 24 h, NS398 increased the production of NS1-associated protein 1, PIBF1 gene, CD24 antigen, and some ESTS. Simultaneously NS398 can inhibit the expression of some oncogenes such as alpha-1-antichymotrypsin, LCN2 (oncogene 24p3), ICAM5, JUN and REL. It was reported that NS398 downregulates the expression of COX-2, nuclear factor-kappaB, p50 and Rel A p65, and its anti-tumor effect is associated with COX-2 transcription inhibition[14].
After being treated for 48 h with NS398, the metastasis-associated genes expressed by LoVo cells changed more profoundly. Thirty-one genes were upregulated, including NS1-associated protein 1, IGFBP5, spectrin, nucleotide binding protein 2, alpha-1-antichymotrypsin, apoptotic protease activating factor, LAMP2, CLCA2, tumor-associated antigen L6, putative insulin-like growth factor, chromosome-associated polypeptide-c, HOXA1, WNT2, FIGF, N-cadherin, PAI2, macrophage-stimulating 1 receptor (MST1R), TIMP1, tachykinin, and precursor.
E-cadherin and N-cadherin are members of the cadherin family of calcium-dependent cell adhesion molecules that play an important role in the embryogenic development and maintenance of normal tissues. N-cadherin present in the most invasive and dedifferentiated breast cancer cell lines, and its exogenous expression in tumor cells induces a scattered morphology and high motility, invasion, and metastasis. N-cadherin co-operates with the fibroblast growth factor receptor, resulting in signals that lead to the upmodulation of MMP-9 and cellular invasion. N-cadherin probably also supports the systemic dissemination of tumor cells by enabling the circulation of tumor cells to associate with the stroma and the endothelium at distant sites[15]. Ectopically expressed N-cadherin fails to assemble cadherin/catenin adhesion complexes and to inhibit invasion. The association of N-cadherin with long P120 (ctn) and tyrosine may explain why N-cadherin cannot replace E-cadherin in pancreatic carcinoma cells[16]. But there are conflicting results of the expression of N-cadherin with the invasion of tumor. P-cadherin is detectable in 40%, N-cadherin in 30%, and E-cadherin in 81% invasive carcinomas. P-cadherin but not E/N-cadherin expression in breast carcinomas shows a strong correlation with higher grade (poorer differentiation), lack of ERs, and presence of EGFR, and its expression may aid in the further subdivision of high grade carcinomas[17]. In the upregulated genes, IGFBP-5 is of great importance. Until now the role of IGFBP-5 in the tumor development is different in different tumors. Expression of IGFBP-5, both by stable transfection and adenoviral-mediated infection, can inhibit the growth of MDA-MB-231 and Hs578T human breast cancer cells over a 13-d period. IGFBP-5 is a potent growth inhibitor and proapoptotic agent in human breast cancer cells via modulation of cell cycle and apoptotic mediators[18].
IGFBP5 is also overexpressed in thyroid tumors. IGFBP-5 and gene 44 are significantly overexpressed in papillary carcinoma[19]. IGFBP-5 mRNA levels are the highest in the benign group without edema of meningiomas, whereas IGFBP-6 mRNA levels are the highest in the group with brain invasion[20]. The presence of IGFBP-5 significantly inhibits cell death induced by C2 or RGD. IGFBP-5 promotes the attachment and survival of Hs578T cells by modulating the balance between ceramide and opposing survival signals[21]. IGFBP-5 has no effect on the proliferation, migration and invasiveness of RSVT2/C cells in vitro[22].
Insulin-like growth factor (IGF)-I and -II are potent mitogens, and can exert autocrine and paracrine effects on growth regulation in human gastric cancer. Their mitogenic effects are regulated by the IGFBPs. The expression pattern of IGFBPs was heterogeneous in the gastric cancer cell lines. IGFBP-2 is expressed in all gastric cancer cell lines, whereas IGFBP-1 is not detectable in any cell line. IGFBP-4 is expressed in most cell lines. IGFBP-3, IGFBP-5, and IGFBP-6 are expressed in approximately 50% of cell lines. In addition, exogenous IGF-I and -II stimulate the proliferation of gastric cancer cells, suggesting the existence of a functional IGF system in gastric cancer. Our data suggest that the IGF-IGFBP system may play an important role in the initiation, progression, and metastasis of gastric cancer[23].
Osteosarcoma cells transfected with IGFBP-5 reduce proliferation under both anchorage-dependent and -independent manner. The increase of proliferation observed in IGFBP-5-secreting clones after addition of exogenous IGF is significantly less than that observed in mock-transfected cells or parental cells. A similar result has been obtained with long [R3] IGF-I which has a low affinity for all IGFBPs, suggesting that the inhibitory effect of IGFBP-5 is only partially IGF-dependent and this effect may be due to an induction of differentiation in these cells because IGFBP-5 increases the normal component secretion of osteosarcoma cells[24].
Upregulation of PAI-2 in LoVo cells may be another mechanism of NS398 underlying the inhibitory effects of tumor cells. PAI-2 is downregulated in esophageal adenocarcinoma compared to normal esophageal tissues[25].
Plasminogen activator inhibitor-2 (PAI-2), a gene whose expression has been linked to cell invasion, has been identified in head and neck tumor cell line. In addition, immunohistochemical evaluation of biopsy samples reveals a high expression of PAI-2 in both normal and dysplastic epithelia with a marked decrease of expression in areas of the biopsies containing HNSCC[26].
The downregulated genes include Max-interacting protein, spectrin, beta-2-microglobulin, CD58 antigen, JUN, APS, BRCA2, CASP1, and some ESTS. BRCA1 and BRCA2 staining increases in the apical cell pole of epithelial malignant cells and in colorectal tumor specimens. Increased BRCA1 and BRCA2 expression may be explained by the fact that colorectal tissue is subjected to very active proliferation and differentiation[27]. High BRCA2 mRNA level is associated with poor outcome and correlates positively and strongly with cell proliferation in breast cancer[28]. NS398 may inhibit the growth of colorectal cancer by downregulating the expression of BRCA2.
In conclusion, the upregulated and downregulated genes identified by cDNA microarray may be the new target genes of NS398.
| 1. | Li M, Wu X, Xu XC. Induction of apoptosis in colon cancer cells by cyclooxygenase-2 inhibitor NS398 through a cytochrome c-dependent pathway. Clin Cancer Res. 2001;7:1010-1016. [PubMed] |
| 2. | Hung WC, Chang HC, Pan MR, Lee TH, Chuang LY. Induction of p27(KIP1) as a mechanism underlying NS398-induced growth inhibition in human lung cancer cells. Mol Pharmacol. 2000;58:1398-1403. [PubMed] |
| 3. | Liu XH, Kirschenbaum A, Yao S, Stearns ME, Holland JF, Claffey K, Levine AC. Upregulation of vascular endothelial growth factor by cobalt chloride-simulated hypoxia is mediated by persistent induction of cyclooxygenase-2 in a metastatic human prostate cancer cell line. Clin Exp Metastasis. 1999;17:687-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 116] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 4. | Attiga FA, Fernandez PM, Weeraratna AT, Manyak MJ, Patierno SR. Inhibitors of prostaglandin synthesis inhibit human prostate tumor cell invasiveness and reduce the release of matrix metalloproteinases. Cancer Res. 2000;60:4629-4637. [PubMed] |
| 5. | Pan MR, Chuang LY, Hung WC. Non-steroidal anti-inflammatory drugs inhibit matrix metalloproteinase-2 expression via repression of transcription in lung cancer cells. FEBS Lett. 2001;508:365-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 6. | Liu LT, Chang HC, Chiang LC, Hung WC. Induction of RECK by nonsteroidal anti-inflammatory drugs in lung cancer cells. Oncogene. 2002;21:8347-8350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 7. | DNA precipitations/Preparation of DNA samples. [Last Update, December1999] [Bioinformatics Manual]. Available from: http: //cmgm.stanford.edu/pbrown/protocols/2_DNA.html. |
| 8. | Post-processing of arrays/Experimental Protocols [updated September 1999] [BioinformaticsManual]. Available from: http: //www.cmgm.stanford.edu/pbrown/protocols/3_post_process.html. |
| 9. | Hasseman JP. Aminoallyl labeling of RNA for microarray. Revision Level: 2. Available from: http: //pga.tigr.org/sop/Moo4_1a.pdf. |
| 10. | Yang IV, Chen E, Hasseman JP, Liang W, Frank BC, Wang S, Sharov V, Saeed AI, White J, Li J. Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol. 2002;3:research0062. [PubMed] |
| 11. | Bushman TL, Kuemmerle JF. IGFBP-3 and IGFBP-5 production by human intestinal muscle: reciprocal regulation by endogenous TGF-beta1. Am J Physiol. 1998;275:G1282-G1290. [PubMed] |
| 12. | Amirghahari N, Harrison L, Smith M, Rong X, Naumann I, Ampil F, Shi R, Glass J, Nathan CO. NS 398 radiosensitizes an HNSCC cell line by possibly inhibiting radiation-induced expression of COX-2. Int J Radiat Oncol Biol Phys. 2003;57:1405-1412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 13. | Peng JP, Liu LT, Chang HC, Hung WC. Enhancement of chemotherapeutic drug-induced apoptosis by a cyclooxygenase-2 inhibitor in hypopharyngeal carcinoma cells. Cancer Lett. 2003;201:157-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 14. | Wen B, Deutsch E, Eschwege P, De Crevoisier R, Nasr E, Eschwege F, Bourhis J. Cyclooxygenase-2 inhibitor NS398 enhances antitumor effect of irradiation on hormone refractory human prostate carcinoma cells. J Urol. 2003;170:2036-2039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | Hazan RB, Qiao R, Keren R, Badano I, Suyama K. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004;1014:155-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 450] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 16. | Seidel B, Braeg S, Adler G, Wedlich D, Menke A. E- and N-cadherin differ with respect to their associated p120ctn isoforms and their ability to suppress invasive growth in pancreatic cancer cells. Oncogene. 2004;23:5532-5542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 64] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 17. | Kovács A, Dhillon J, Walker RA. Expression of P-cadherin, but not E-cadherin or N-cadherin, relates to pathological and functional differentiation of breast carcinomas. Mol Pathol. 2003;56:318-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 18. | Butt AJ, Dickson KA, McDougall F, Baxter RC. Insulin-like growth factor-binding protein-5 inhibits the growth of human breast cancer cells in vitro and in vivo. J Biol Chem. 2003;278:29676-29685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 117] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 19. | Stolf BS, Carvalho AF, Martins WK, Runza FB, Brun M, Hirata R, Jordão Neves E, Soares FA, Postigo-Dias J, Kowalski LP. Differential expression of IGFBP-5 and two human ESTs in thyroid glands with goiter, adenoma and papillary or follicular carcinomas. Cancer Lett. 2003;191:193-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 20. | Nordqvist AC, Mathiesen T. Expression of IGF-II, IGFBP-2, -5, and -6 in meningiomas with different brain invasiveness. J Neurooncol. 2002;57:19-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 21. | McCaig C, Perks CM, Holly JM. Signalling pathways involved in the direct effects of IGFBP-5 on breast epithelial cell attachment and survival. J Cell Biochem. 2002;84:784-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (1)] |
| 22. | Lee BP, Rushlow WJ, Chakraborty C, Lala PK. Differential gene expression in premalignant human trophoblast: role of IGFBP-5. Int J Cancer. 2001;94:674-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 23. | Yi HK, Hwang PH, Yang DH, Kang CW, Lee DY. Expression of the insulin-like growth factors (IGFs) and the IGF-binding proteins (IGFBPs) in human gastric cancer cells. Eur J Cancer. 2001;37:2257-2263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 76] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Schneider MR, Zhou R, Hoeflich A, Krebs O, Schmidt J, Mohan S, Wolf E, Lahm H. Insulin-like growth factor-binding protein-5 inhibits growth and induces differentiation of mouse osteosarcoma cells. Biochem Biophys Res Commun. 2001;288:435-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 25. | Hourihan RN, O'Sullivan GC, Morgan JG. Transcriptional gene expression profiles of oesophageal adenocarcinoma and normal oesophageal tissues. Anticancer Res. 2003;23:161-165. [PubMed] |
| 26. | Hasina R, Hulett K, Bicciato S, Di Bello C, Petruzzelli GJ, Lingen MW. Plasminogen activator inhibitor-2: a molecular biomarker for head and neck cancer progression. Cancer Res. 2003;63:555-559. [PubMed] |
| 27. | Bernard-Gallon DJ, Peffault de Latour M, Hizel C, Vissac C, Cure H, Pezet D, Dechelotte PJ, Chipponi J, Chassagne J, Bignon YJ. Localization of human BRCA1 and BRCA2 in non-inherited colorectal carcinomas and matched normal mucosas. Anticancer Res. 2001;21:2011-2020. [PubMed] |
| 28. | Bièche I, Tozlu S, Girault I, Lidereau R. Identification of a three-gene expression signature of poor-prognosis breast carcinoma. Mol Cancer. 2004;3:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
Science Editor Wang XL and Li WZ Language Editor Elsevier HK