Published online Jun 14, 2005. doi: 10.3748/wjg.v11.i22.3473
Revised: July 13, 2004
Accepted: August 30, 2004
Published online: June 14, 2005
AIM: To identify the pre-X region in hepatitis B virus (HBV) genome and to study the relationship between the genotype and the pre-X region. To investigate the biological function of whole-X (pre-X plus X) protein, we performed yeast two-hybrid to screen proteins in liver interacting with whole-X protein.
METHODS: The pre-X region of HBV was amplified by polymerase chain reaction (PCR) method, and was cloned to pGEM Teasy vector. After the target region was sequenced, Vector 8.0 software was used to analyze the sequences. The whole-X bait plasmid was constructed by using yeast two-hybrid system 3. Yeast strain AH109 was transformed. After expression of the whole-X protein in AH109 yeast strains was proved, yeast two-hybrid screening was performed by mating AH109 with Y187 containing liver cDNA library plasmid. The mated yeast was plated on quadruple dropout medium and assayed for α-gal activity. The interaction between whole-X protein and the protein obtained from positive colonies was further confirmed by repeating yeast two-hybrid. After extracting and sequencing of plasmid from blue colonies, we carried out analysis by bioinformatics.
RESULTS: After sequencing, 27 of 45 clones (60%) were found encoding the pre-X peptide. Eighteen of twenty-seven clones (66.7%) of pre-X coding sequences were found from genotype C. Five positive colonies that interacted with whole-X protein were obtained and sequenced; namely, fetuin B, UDP glycosyltransferase 1 family-polypeptide A9, mannose-P-dolichol utilization defect 1, fibrinogen-B beta polypeptide, transmembrane 4 superfamily member 4-CD81 (TM4SF4).
CONCLUSION: The pre-X gene exists in HBV genome. Genes of proteins interacting with whole-X protein in hepatocytes were successfully cloned. These results brought some new clues for studying the biological functions of whole-X protein.
- Citation: Yang Q, Cheng J, Dong J, Zhang J, Zhang SL. Molecular epidemiological study on pre-X region of hepatitis B virus and identification of hepatocyte proteins interacting with whole-X protein by yeast two-hybrid. World J Gastroenterol 2005; 11(22): 3473-3478
- URL: https://www.wjgnet.com/1007-9327/full/v11/i22/3473.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i22.3473
Hepatitis B virus (HBV) predominantly infects host hepatocytes and causes a spectrum of pathological processes, ranging from occult infection to the later development of primary liver cancer[1,2]. HBV genomes have a compact genetic organization with four open reading frames (ORFs) as P, X, S, and C corresponding to the polymerase, the X protein, the S including pre-S, and the nucleocapsid/HBeAg proteins, respectively. The P gene covers more than 70% of the complete genome and overlaps the entire pre-S and S-genes and the X and core genes partially[3,4]. pX is likely to be an important regulatory protein since its sequence is conserved among the mammalian hepadnaviridae members. pX increases HBV transcription by trans-activating the viral enhancer-I via the sequence named the E-element. HBx deregulates cell cycle checkpoints and stimulates DNA synthesis, leading to the proliferation of quiescent fibroblasts. Importantly, HBx is a moderate but broad-acting transcriptional transactivator and activates a variety of cellular and viral genes, including proto-oncogenes, such as c-myc, c-fos, and c-jun. However, little is known about the exact role of HBx in tumorigenesis[5-7].
Some of the reported HBV sequences have an additional in-frame initiation codon, ATG, 56 nucleotide triplets upstream of X gene’s ATG. In our study on molecular epidemiological features of HBV genome, the ORF named pre-X region was found[8,9]. The pre-X gene can be translated with X gene in frame. The pre-X plus X gene, named whole-X gene, is 630 bp long and the whole-X polypeptide encoded by this gene comprises 210 amino acids. In order to further reveal the biological roles of whole-X protein, we therefore tried to identify its associated proteins in the present study by the GAL4-based yeast two-hybrid system using the full-length whole-X cDNA as a bait to screen the human fetal liver cDNA library.
Matchmaker GAL4 two-hybrid system 3 and vector pACT2 containing the human liver cDNA library were obtained from Clontech Co., USA. Yeast strain AH109 (MATa, trp 1-901, leu2-3, 112, ura 3-52, his3-200, gal4, gal80, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2 URA3::MEL1TATA-lacZ MEL1) contains pGBKT7-53, coding for DNA-BD/mouse p53 fusing protein. Yeast strain Y187Y187 (MATa ura3-52, his3-200, Ade2-101, trp1-901, leu2-3, 112, gal4, gal80, met-, URA3::GAL1UAS-GAL1TATA-lacZ MEL1) contains pTD1-1, in which pACT2 coding for AD/SV40 large T antigen fusing protein. AH109 was used for cloning of bait plasmids and Y187 was used for cloning of library plasmids. Yeast-Escherichia coli shuttle plasmids, pGBKT7 DNA-BD cloning plasmid, pGADT7 AD cloning plasmid, pGBKT7-53 control plasmid, pGADT7, pGBKT7-Lam control plasmid, pCL1 plasmid were obtained from Clontech Ltd Co. (K1612-1).
Taq DNA polymerase was purchased from Promega Co. T4 DNA ligase, EcoRI and BamHI restriction endonuclease were purchased from Takara Co. c-Myc mAb secreted by 1-9E10.2 hybridoma (ATCC), goat anti-mouse IgG conjugated with horseradish peroxidase were from Zhongshan Company, China. Tryptone and yeast extracts from OXOID. X-a-Gal and cultural media: YPDA, SD/-Trp SD/-Leu, SD/-Trp/-Leu, SD/-Trp/-Leu/-His, SD/-Trp/-Leu/-His/-Ade from Clontech Ltd Co.
DNA was extracted from serum of 17 patients with chronic hepatitis B. The extracted DNA was mixed with polymerase chain reaction (PCR) mixture containing the primers: sense 5’- CCA AGT GTT TGC TGA CGC AAC C’, antisense 5’- GGA TCC AGT TGG CAG CAC ACC-3’. The PCR product was cloned with pGEM-T vector (Promega Co.). The primary structure of insert was confirmed by direct sequencing[10,11].
The full-length whole-X gene of HBV was cloned into the yeast two-hybrid BD vector using a BamHI and EcoRI, which could facilitate expression of DNA binding domain, c-myc and whole-X fusion protein. The construction was verified by restriction digestion and sequencing (Figure 1).
Denatured cell extracts were subjected to electrophoresis using 4-12% SDS-polyacrylamide gels and transferred to nitrocellulose membranes[14]. The membranes were blocked with 5% non-fat dry milk for 1 h and then incubated with monoclonal anti-c-myc antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA) for 2 h and with HRP-conjugated secondary antibody for another 1 h prior to detection of antibody reactive proteins with chemiluminescence reagent (ECL, Amersham Pharmacia Biotech).
The screening protocol was detailed previously[12]. Yeast transformants were plated and selected on media lacking leucine, tryptophan, histidine, and adenine. After 6-18 d, the yeast colonies were transferred onto the plates containing X-a-Gal to check for expression of the MEL1 reporter gene (blue colonies).
Yeast plasmid was isolated with the lyticase method (provided by Clontech Co.) and transformed into E. coli strain DH5α and amplified in bacteria. Plasmids isolated from bacteria were reintroduced into yeast strain Y187, then mating experiments were carried out by mating with yeast strain AH109 containing pGBKT7-whole-X or pGBKT7-Lam. The diploid yeast was plated on media lacking leucine, tryptophan, histidine, and adenine with X-α-Gal.
After the positive colonies were sequenced, the sequences were blasted with GenBank to analyze the function of the genes.
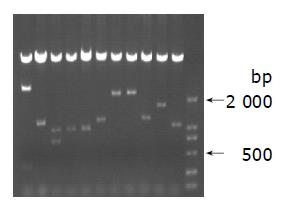
After sequencing, 27 of 45 clones (60%) were found encoding the pre-X peptide. Eighteen of twenty-seven clones (66.7%) of pre-X coding sequences were found from genotype C. Three types of replacement mutation led to pre-mature coding of pre-X gene. The mutation in pre-X peptide had the feature of individual mutation (Figure 1).
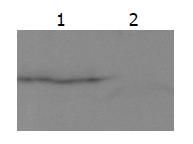
Yeast strain AH109 transformed with pGBKT7-whole-X could stably express the fusion protein at higher level and could only grow on SD/-Trp medium (Figure 2).
The whole-X protein was used as the bait for screening a human liver yeast two-hybrid library. One hundred and twenty-six clones grew in the absence of tryptophan, leucine, histidine, adenine. The clones were processed for β-galactosidase assay, and blue colonies were picked. pACT2/cDNA plasmids were isolated and eliminated by EcoRI digestion (Figure 3). After elimination, 14 positive clones were further tested for specificity of β-galactosidase expression (Figure 4). After confirmation of the true interaction with whole-X protein in yeast, five independent positive clones were identified and sequenced (Figure 4).
The nucleotide sequences of five clones from this cDNA library were analyzed[24], the full length sequences were obtained with Vector NTI 6 and by searching BLAST database (http://www.ncbi.nlm.nih.gov/). Some genes coding proteins were involved in cell cycle regulation, and apoptosis, signal transduction pathway and tumor development (Table 1).
| Known genes | Number of clones (%) | Homologous |
| Fetuin B | 4 | 100 |
| UDP glycosyltransferase 1 family, polypeptide A9 | 2 | 99 |
| Mannose-P-dolichol utilization defect 1 (MPDU1) | 3 | 100 |
| Fibrinogen, B beta polypeptide (FGB) | 2 | 100 |
| Transmembrane 4 superfamily member 4 (TM4SF4) | 2 | 100 |
HBV belongs to the family of hepadnaviruses. It has a 3200 bp partially double-stranded DNA genome from which four major classes of transcripts are synthesized. The 3.5 kb pregenomic RNA not only serves as template for reverse transcription, but also contains coding regions for nucleocapsid protein and reverse transcriptase. A subclass of this transcript with a slightly longer 59 end codes for the precore protein, which, after processing, is secreted as HBV e antigen (HBeAg). The 2.4 kb RNA encompasses the preS1 ORF that encodes the large surface (L) protein. The 2.1 kb RNA contains the preS2 and S ORFs that encode the middle (M) and small (s) surface proteins, respectively. The smallest transcript (approximately 0.9 kb) codes for the X protein. Mutations and deletions in the HBV genome have frequently been detected during persistent viral infection[13,14].
Takahashi et al[15], in an infected serum, have identified a protein that is coded for by the X gene of HBV, and found a polypeptide that was weakly bound with an anti-X-mAb. This result is hard to re-confirm with different sera. They sequenced the upstream region of the gene of HBV DNA from the initial sample and found that there is a pre-X ORF of 56 codons, the pre-X variant is frequently detected in chronic liver disease, but seldom in asymptomatic carriers. Another characteristic of the pre-X variant is the cancellation of in-frame stop codon, suggesting that pre-X and X are linearly translated in this variant. Loncarevic et al[17], reported an HBV genome that possesses an intact pre-X ORF.
In our study, 17 samples were collected. One was genotype A, 3 were genotype B, 10 were genotype C and 3 were B/C genotype mixture. After sequencing, 27 of 45 clones (60%) were found encoding the pre-X peptide. Eighteen of twenty-seven clones (66.7%) pre-X coding sequences were found from genotype C. Three types of replacement mutation lead to pre-mature coding of pre-X gene. The mutation in pre-X peptide had the feature of individual mutation, suggesting that coding of the pre-X gene is popular in HBV genome.
The pre-X mutations may have some effect on expression of the X gene. Loncarevic et al[17], reported that all the five HBV DNA clones derived from hepatocellular carcinoma had intact pre-X ORF. Protein–protein interactions play an important role in almost all events that takes place in cells. The two-hybrid screen is a promising experimental approach to identify whole-X protein interacting proteins.
The yeast two-hybrid system is a better choice of detecting protein-protein interactions. Yeast two-hybrid system 3 based on the system is commercially available from Clontech Co.[18-20]. In this system, the promoters controlling HIS3, ADE2, and MEL1 expression in AH109 have significantly fewer false positives and the simple mating protocol significantly reduces the labor and time involved in performing a two-hybrid library screening and improves the chances of finding rare protein-protein interactions and leads to more reproducible results[21,22]. On screening a human liver cDNA library, five putative clones are identified as associated proteins, namely fetuin B, UDP glycosyltransferase 1 family-polypeptide A9, MPDU1, FGB, transmembrane 4 superfamily member 4-CD81 (TM4SF4). Mammalian transmembrane 4 superfamily (TM4SF) proteins (also known as tetraspans or tetraspanins) include at least 16 core members and a number of additional proteins with sequence similarities. Almost all mammalian cells contain one or more TM4SF proteins[23]. TM4SF protein CD81 may function in cell migration, proliferation and tumor cell metastasis. Most TM4SF proteins can be found on plasma membrane, and several are located in cell lamellipodia and lopodia, consistent with their role in cell motility. TM4SF proteins including CD81 are also found in various intracellular granules and vesicles. A specific subset of TM4SF proteins may recruit PI 4-kinase to specific membrane locations, and thereby influence phosphoinositide-dependent signaling[24]. Fetuin B is a liver-produced negative acute phase protein. Multiple physiological roles of the protein have been suggested, including ability to bind to hydroxyapatite crystals and to specifically inhibit the tyrosine kinase activity of the insulin receptor. It is mainly a fetal protein, in the sense that the highest concentrations are found in serum and body fluids of embryos and fetuses[25]. Fetuin might be involved in cell differentiation and tissue transformation during the initial histogenesis[26]. The result of this study shows that the whole-X protein may modulate signal transduction pathway by protein-protein binding and carcinoma formation.
| 1. | Bertoletti A, Ferrari C. Kinetics of the immune response during HBV and HCV infection. Hepatology. 2003;38:4-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 186] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 2. | Besisik F, Karaca C, Akyüz F, Horosanli S, Onel D, Badur S, Sever MS, Danalioglu A, Demir K, Kaymakoglu S. Occult HBV infection and YMDD variants in hemodialysis patients with chronic HCV infection. J Hepatol. 2003;38:506-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 63] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 3. | Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H. Structural organization of the hepatitis B virus minichromosome. J Mol Biol. 2001;307:183-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 288] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 4. | Sterneck M, Kalinina T, Günther S, Fischer L, Santantonio T, Greten H, Will H. Functional analysis of HBV genomes from patients with fulminant hepatitis. Hepatology. 1998;28:1390-1397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 42] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 5. | Kim KH, Seong BL. Pro-apoptotic function of HBV X protein is mediated by interaction with c-FLIP and enhancement of death-inducing signal. EMBO J. 2003;22:2104-2116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 110] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 6. | Lee MO, Choi YH, Shin EC, Kang HJ, Kim YM, Jeong SY, Seong JK, Yu DY, Cho H, Park JH. Hepatitis B virus X protein induced expression of interleukin 18 (IL-18): a potential mechanism for liver injury caused by hepatitis B virus (HBV) infection. J Hepatol. 2002;37:380-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 58] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 7. | Hoare J, Henkler F, Dowling JJ, Errington W, Goldin RD, Fish D, McGarvey MJ. Subcellular localisation of the X protein in HBV infected hepatocytes. J Med Virol. 2001;64:419-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 8. | Dong J, Cheng J. Study on definition of pre-X region in hepatitis B virus genome. Shijie Huaren Xiaohua Zazhi. 2003;11:1097-1101. |
| 9. | Dong J, Cheng J, Wang QH, Liu Y, Wang G, Shi SS, Xia XB, Shao Q, Si CW. The preliminary study on hepatitis B virus (HBV) quasispecies in patients with chronic HBV infection. Zhonghua Chuanranbing Zazhi. 2001;19:199-203. |
| 10. | Yang Q, Dong J, Cheng J, Liu Y, Hong Y. Definition of pre-X promoter sequence from hepatitis B virus genome and characterization of it’s transcription activity. JiefangJun YiXue Zazhi. 2003;28:763-765. |
| 11. | Li K, Wang L, Cheng J, Lu YY, Zhang LX, Li L, Liu Y, Duan HJ. Expression of NS2 gene of hepatitis C virus from yeast two hybrid’Bait’ vector in yeast. Shijie Huaren Xiaohua Zazhi. 2002;10:129-132. |
| 12. | Yang Q, Cheng J, Liu Y, Hong Y, Wang JJ, Zhang SL. Cloning and analysis of genes transactivated by NS5A-TP4 protein by suppression subtractive hybridization. Zhongxiyi Jiehe Ganbing Zazhi. 2003;13:325-335. |
| 13. | Locarnini S. Molecular virology of hepatitis B virus. Semin Liver Dis. 2004;24 Suppl 1:3-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 136] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 14. | Arad U, Axelrod J, Ben-nun-Shaul O, Oppenheim A, Galun E. Hepatitis B virus enhances transduction of human hepatocytes by SV40-based vectors. J Hepatol. 2004;40:520-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 15. | Takahashi K, Kishimoto S, Ohori K, Yoshizawa H, Akahane Y, Okamoto H, Mishiro S. A unique set of mutations in the “preX” region of hepatitis B virus DNA frequently found in patients but not in asymptomatic carriers: implication for a novel variant. Int Hepatol Commun. 1995;3:131-138. |
| 16. | Takahashi K, Akahane Y, Hino K, Ohta Y, Mishiro S. Hepatitis B virus genomic sequence in the circulation of hepatocellular carcinoma patients: comparative analysis of 40 full-length isolates. Arch Virol. 1998;143:2313-2326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 130] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 17. | Loncarevic IF, Zentgraf H, Schröder CH. Sequence of a replication competent hepatitis B virus genome with a preX open reading frame. Nucleic Acids Res. 1990;18:4940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 18. | Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4257] [Cited by in RCA: 4357] [Article Influence: 117.8] [Reference Citation Analysis (5)] |
| 19. | Evangelista C, Lockshon D, Fields S. The yeast two-hybrid system: prospects for protein linkage maps. Trends Cell Biol. 1996;6:196-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 39] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 20. | Kharel Y, Takahashi S, Yamashita S, Koyama T. In vivo interaction between the human dehydrodolichyl diphosphate synthase and the Niemann-Pick C2 protein revealed by a yeast two-hybrid system. Biochem Biophys Res Commun. 2004;318:198-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 21. | Vidalain PO, Boxem M, Ge H, Li S, Vidal M. Increasing specificity in high-throughput yeast two-hybrid experiments. Methods. 2004;32:363-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 110] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 22. | von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1668] [Cited by in RCA: 1421] [Article Influence: 59.2] [Reference Citation Analysis (0)] |
| 23. | Fan J, Hooker CW, McManus DP, Brindley PJ. A new member of the transmembrane 4 superfamily (TM4SF) of proteins from schistosomes, expressed by larval and adult Schistosoma japonicum. Biochim Biophys Acta. 1997;1329:18-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 24. | Hashida H, Takabayashi A, Tokuhara T, Hattori N, Taki T, Hasegawa H, Satoh S, Kobayashi N, Yamaoka Y, Miyake M. Clinical significance of transmembrane 4 superfamily in colon cancer. Br J Cancer. 2003;89:158-167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 101] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 25. | Denecke B, Gräber S, Schäfer C, Heiss A, Wöltje M, Jahnen-Dechent W. Tissue distribution and activity testing suggest a similar but not identical function of fetuin-B and fetuin-A. Biochem J. 2003;376:135-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 229] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 26. | Leite-Browning ML, McCawley LJ, Choi OH, Matrisian LM, Ochieng J. Interactions of alpha2-HS-glycoprotein (fetuin) with MMP-3 and murine squamous cell carcinoma cells. Int J Oncol. 2002;21:965-971. [PubMed] |