Published online Jan 14, 2005. doi: 10.3748/wjg.v11.i2.193
Revised: April 8, 2004
Accepted: May 24, 2004
Published online: January 14, 2005
AIM: To evaluate the effects of survivin on cell proliferation and apoptosis in liver cancer.
METHODS: MTT assay was used to generate and optimize phosphorothioate antisense oligonucleotides (ODNs)-LipofectamineTM2000 (LiP) compound by varying ODNs (μg):LiP (μL) ratios from 1:0.5 to 1:5. Then, liver cancer cells (HepG2) were transfected with the compound. By using RT-PCR and Western blot, the expression levels of survivin mRNA and proteins were detected in HepG2 cells treated with antisense compounds (ODNs:LiP = 1:4), and compared with those treated with sense compounds (1:4) as control. MTT assay was applied to the determination of cell proliferation in HepG2 cells. Active caspase-3 was evaluated by flow cytometric analysis. The morphological changes were assessed by electron microscopy. Laser scanning confocal microscopy was performed to detect the subcellular localization of survivin proteins in treated and untreated cells.
RESULTS: Antisense compounds (1:4) down-regulated survivin expression (mRNA and protein) in a dose-dependent manner with an IC50 of 250 nmol/L. Its maximum effect was achieved at a concentration of 500 nmol/L, at which mRNA and protein levels were down-regulated by 80%. The similar results were found in MTT assay. Antisense compound (1:4)-treated cells revealed increased caspase-3-like protease activity compared with untreated cells. Untreated cells as control were primarily negative for the presence of active-caspase-3. As shown by transmission electron microscopy, treated cells with antisense compounds (1:4) resulted in morphological changes such as blebbing and loss of microvilli, vacuolization in the cytoplasm, condensation of the cytoplasm and nuclei, and fragmented chromatin. Immunofluorescence analysis confirmed the presence of survivin protein pool inside the cytoplasm in untreated cells. Labeled-FITC immunofluorescence staining of survivin clearly showed that survivin was distributed mainly in a spotted form inside the cytoplasm. Whereas cells treated with antisense compounds were rare and weak inside the cytoplasm.
CONCLUSION: Down-regulation of survivin expression induced by the antisense compounds reduces tumor growth potential, promotes apoptosis and affects the localization of survivin proteins in HepG2 cells. Furthermore, survivin protein is a key molecule associated with proliferation and apoptosis, and antisense oligonucleotides targeting survivin have a bright prospect in the therapy of liver cancer.
- Citation: Dai DJ, Lu CD, Lai RY, Guo JM, Meng H, Chen WS, Gu J. Survivin antisense compound inhibits proliferation and promotes apoptosis in liver cancer cells. World J Gastroenterol 2005; 11(2): 193-199
- URL: https://www.wjgnet.com/1007-9327/full/v11/i2/193.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i2.193
Diminished apoptosis plays a critical role in tumor initiation, progression, as well as in cancer therapy[1]. Several proteins that inhibit apoptosis have been identified, including bcl-2 family members bcl-2, bcl-xl and IAP[2]. Certain members of the latter family directly inhibit terminal effector caspases engaged in the execution of cell death[3]. Among molecules of the IAP family, survivin is unique in having a single baculovirus IAP repeat (BIR) and extended C-terminal x-helix[4] and dimeric architecture that is essential for the inhibition of apoptosis [5,6]. Although survivin protein lacks the ability to directly inhibit caspase-3[7], it binds quantitatively to a new IAP-inhibiting protein, Smac/Diablo[8,9], raising the possibility that it might suppress caspases indirectly by freeing other IAP family members from the constraints of protein. Taken together, these studies support the notion that survivin exerts an anti-apoptotic effect. Survivin is expressed during embryonal development but lacks expression in terminally differentiated adult tissues[2,4]. Interestingly, it becomes reexpressed in transformed cell lines and in a variety of human tumors[3,4]. Survivin is expressed in the G2/M phase of the cell cycle and associates with microtubules of the mitotic spindle in a specific and saturable reaction that is regulated by microtubule dynamics[10]. This implies that overexpression of survivin has oncogenic potential because it may overcome the G2/M phase checkpoint to enforce progression of cells through mitosis, thus promoting proliferation. Recent studies suggest an alternative distribution of survivin localized to kinetochores of metaphase chromosomes, and to microtubules of the central spindle midzone at anaphase[11-13]. Approximately 80% of the total cellular survivin content in mitotic cells is bound to centrosomes and microtubules of the metaphase and anaphase spindles[10]. These studies reveal that survivin could connect the cell cycle with apoptosis, thus providing a death switch for the termination of defective mitosis. These lines of evidence make survivin an attractive therapeutic target in cancer treatment.
Liver cancer is a leading cause of cancer death, and its incidence continues to rise. The main reasons for the unfavorable prognosis of these tumors are their propensity to metastasizing early and developing resistance to a wide range of functionally anticancer agents. Recent studies show that expression of survivin is not only detected in stomach cancer[14,15], non-small cell lung cancer[16], breast cancer[17], esophageal cancer[18], colon cancer, ovarian carcinoma[19], but also in HepG2, Huh7, sk-Hep1 cell lines and hepatocellular carcinoma tissues, and it has a close correlation with apoptosis and proliferation of liver cancer[4,20]. In addition, skin cancer HaCat cells transfected with antisense oligonucleotide down regulate expression of survivin, promote apoptosis and diminish proliferation[21]. Consequently, targeting expression of survivin has a potential value in cancer therapy. Although antisense oligonucleotide has been widely recognized as an efficient tool for the inhibition of gene expression in a sequence specific way, during practical application of antisense approach, many key problems need to be solved, such as to find an effective targeting site of survivin mRNA that is likely to be accessible to antisense oligonucleotides, to select an adaptable vector combined with antisense oligonucleotides in the formation of a potent compound to facilitate apoptosis of tumor, to optimize transfection conditions in a cell line with high expression of survivin.
In the present study, we generated a series of antisense compounds, with the strongest efficient antisense oligonucleotides designed by Olie et al[22] and highly efficient vector Lipofecta-mine 2000. Using MTT assay and the survivin-overexpressing liver cancer cell line HepG2, one antisense compound was identified that could most efficiently down-regulate survivin expression levels (mRNA and protein) and directly induce apoptosis. Furthermore, in a double immunofluorescence staining experiment with FITC-labelled antibody against survivin and Allexa-labelled concanavalin A, evidence is provided that antisense mediated down-regulation of survivin has the potential to affect subcellular localization of survivin proteins in HepG2 cells.
According to the method of Olie et al[22], antisense and sense oligonucleotides for survivin were designed. The antisense oligonucleotides targeting nucleotides 232-251 revealed the strongest effect and were used in the following experiments. Their sequences are shown in Table 1. Phosphorothioate oligonucleotides ODNs were purchased from Sangon (Shanghai, China), and delivered in the form of complex with Lipofectamine 2000 (Lip) (Invitrogen, USA).
| Antisense | 5'-CCCAGCCTTCCAGCTCCTTG-3' |
| Sense | 5'-CAAGGAGCTGGAAGGCTGGG-3' |
Liver cancer cell line HepG2, which was reported to express high levels of survivin[19], was obtained from Shanghai Institute of Cell Biology. Cells were grown in DMEM (Gibco, USA) supplemented with 10% heat-inactivated fetal bovine serum(Sijiqing, Hangzhou, China). Cells were maintained in monolayer cultures at 37 °C in a humidified atmosphere consisting of 5 mL/L CO2 and 95% air.
To obtain the highest transfection efficiency and low non-specific effects, transfection conditions were optimized by varying ODNs, LiP concentrations and cell density. Cells should be greater than 80% confluence and ODNs (μg):LiP (μL) ratios were varied from 1:0.5 to 1:5. ODNs were diluted in the appropriate amount of DMEM without serum, at the final concentration of 0.2 μg/25 μL, and they were mixed gently. LiP was gently mixed before use, then diluted in an appropriate amount of DMEM without serum at the final concentrations of 0.1 μg/25 μL, 0.2 μg/25 μL, 0.4 μg/25 μL, 0.6 μg/25 μL, 0.8 μg/25 μL, 1 μg/25 μL. These ODNs solutions were gently mixed and incubated for 5 min at room temperature. After incubated for 5 min, the diluted ODNs were mixed with the six different concentrations of diluted LiP (1:0.5-1:5), and incubated for 20 min at room temperature to form the ODNs-LiP compounds. Fifty microliters of ODNs-LiP compounds was added to each well (96-well plates) containing cells and media, gently mixed again by rocking the plate back and forth, the cells were incubated at 37 °C in a CO2 incubator for 48 h. Then, MTT assay was performed.
One day before transfection, HepG2 cells were plated in 96-, 24-, or 6-well tissue culture plates. Oligonucleotides were delivered in the form of compound LiP as described above. After a 24-h transfection, the transfection medium was replaced by medium without transfection reagents. HepG2 cells were harvested 24, 48 or 72 h after the start of transfection.
Growth inhibition in HepG2 cell line was determined by a colorimetric MTT assay. HepG2 cells were cultured in 96-well plates with an initial concentration of 1×104 cells/well in DMEM supplemented with 10% fetal bovine serum. ODNs were delivered in the form of compound LiP, as described above. Briefly, after transfection for 48 h at 37 °C, 20 μL of MTT reagent(Sangon, Shanghai, China) was added and allowed to react for 4 h at 37 °C. Subsequently, the MTT was discarded, 150 μL of DMSO reagent (Sangon, Shanghai, China) was added and allowed to react for 15 min at 37 °C. Substrate cleavage was monitored at 570 nm by a microplate reader (Tosoh, Yamaguchi, Japan) and the rate of inhibition was calculated. Experiments were repeated three times.
Total cellular RNA was extracted using RNeasy Plant Mini kit reagent according to the manufacturer’s recommendations(QIAGEN, Germany). The quantity and quality of RNA were assessed spectrophotometrically at 260 nm and 280 nm (the A260 to A280 ratio of pure RNA was approximately 2).
RT-PCR amplification product was synthesized with 0.1 μg of total RNA using OneStep RT-PCR kit (QIAGEN, Germany) according to the manufacturer’s instructions. Primers used for amplification were human survivin sense primer corresponding to nucleotides 47-66 (5’-GGCATGGGTGCCCCGACGTT-3’), and antisense primer complementary to nucleotides 466-485 (5’-AGAGGCCTCAATCCATGGCA-3’). Amplification of human β-actin served as an internal control, β-actin sense primer was corresponding to nucleotides 578-609 (5’-ATCTGGCACCACACCTTCTACAATGAGCTGCG-3’), and antisense primer was complementary to nucleotides 1 415-1 384 (5’- CGTCATACTCCTGCTTCCTGATCCACATCTGC-3’). Optimization conditions were as follows: reverse transcription for 30 min at 50 °C, initial PCR activation step for 15 min at 95 °C. Three-step cycling was denaturation for 1 min at 94 °C, annealing for 1 min at 62 °C,extension for 1 min at 72 °C, and a final extension for 10 min at 72 °C using the Eppendorf PCR System (Eppendorf, Germany). PCR products were analysed on a 2% agarose gel by electrophoresis and the bands were visualized by ethidium bromide staining.
Cells were collected on ice and centrifuged at 1500 rm for 5 min at 4 °C. The cells were resuspended and washed with ice-cold PBS and centrifuged again at 1500 rm for 5 min at 4 °C. Supernatants were discarded and cell pellets were lysed in 2× cell lysis buffer [2% Triton X-100, 300 mmol/L NaCl, 20 mmol/L Tris-Cl pH 7.2, 10% glycerol and 4% SDS] and heated to 95 °C for 10 min. The lysates were centrifuged at 14000 rm for 30 min at 4 °C and the protein supernatants were collected and stored at -20 °C. The protein concentrations were determined by BCA assay according to the manufacturer’s instructions (Pierce Co). Equal quantities of proteins (20 μg/well) were separated on 7.5-12% sodium dodecyl sulfate polyacrylamide gel electrophoretic gels for the detection of survivin. Following electrophoretic transfer of protein onto nitrocellulose membranes, immunoblots were sequentially incubated in 5% skimmed milk blocking solution at room temperature for 2 h. The membranes were incubated in a solution containing antihuman survivin antibody (1:800; Oncogene, USA) overnight at 4 °C. After washed three times with 1% PBS-Tween solution at room temperature for 1 h, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1:2000; Oncogene, USA) at room temperature for 2 h. Finally, after the membranes were washed for 1 h, protein bands were detected by using the ECL system according to the manufacturer’s; instructions (Amersham Life Sciences). β-actin was used as a protein loading control.
Caspase-3-like protease activity in cells was analyzed by flow cytometry (FACS Calibur, Bection Dikinson, USA) using a PE-conjugated polyclonal rabbit anti-active caspase-3 antibody kit (Becton Dickinson, USA). Zero, 24, 48, 72 h after antisense compound induction respectively, cells were harvested and washed twice with PBS, fixed in ice-cold 70% ethanol and stored at 4 °C. Prior to analysis, cells were again washed with PBS. Cytometric analyses were performed using a flow cytometer and Cell Quest software. Approximately 20000 cells were calculated for each determination.
Cells were seeded into 30-mm dishes, treated as described above, collected on ice with media by gently scraping, and washed three times in ice-cold PBS. The cell pellets were fixed with a solution of 2% formaldehyde and 3% glutaraldehyde in 0.1 mol/L sodium cacodylate buffer (pH 7.4) for 1 h at 48 °C. The fixed cells were washed three times in cacodylate buffer (pH 7.2) containing 0.2 mol/L sucrose, postfixed with 1% osmic acid in 0.3 mol/L cacodylate buffer, dehydrated in a graded series of acetone, and embedded in epoxy resin. Ultrathin sections were cut, stained with uranyl acetate and lead citrate, and assessed by using a TECNA10 electron microscope (Philips, Holand).
Cells were cultured on microscope slides, then treated or untreated with antisense compounds (1:4) for 48 h. Cells were washed three times with PBS for 15 min and fixed in ice-cold paraformaldehyde for 30 min. Cells were fixed in ice-cold CAM (chloroform:acetone:methanol = 1:1:2) at -20 °C for 30 min, and washed three times with PBS for 15 min. Cells were incubated overnight at 4 °C with polyclonal anti-survivin (1:400 diluted; rabbit polyclonal IgG; Oncogene, USA), washed three times with PBS for 30 min, incubated for 2 h at room temperature with fluorescein (FITC)-conjugated anti-rabbit-IgG (1:1000; Oncogene, USA). After a further washing step, cell membranes were stained for 2 min with a 5×10-3% (w/v) solution of Alexa-labelled concanavalin A (MoBiTec, Gottingen, Germany) in BPS. Finally, the cells were embedded in a gel containing 22.73% (w/w) glycerine, 9.1% (w/w) Mowiol4-88 (present from Ningbo Institute of Microcirculation and Henbane), 22.73% (w/w) double-distilled water, 45.45% (w/w) 0.2 mol/L Tris buffer, pH 8.5, and 2.5% (w/w) DABCO (present from Ningbo Institute of Microcirculation and Henbane) as an antifading agent. After 12 h, confocal microscopy was performed with a microscope (TCS-SP2, Leica, Germany) equipped with an argon/krypton laser. Two-channer image recording at 488 nm and 633 nm laser excitation was used. Optical filters were chosen for the FITC and TRLTC range. All optical sections were recorded with the same laser and detector settings using software (Leica Confocal, Germany). Further image processing was performed with software (Leica Confocal, Germany) on a computer system. Confocal stacks of green and red fluorescence were visualized in section view mode.
All statistical analyses were performed by the SPSS11.0 software package for Windows (SPSS Inc., Chicago, IL). The t test was used to compare the distribution of individual variables. A two-tailed P value less than 0.05 was considered statistically significant.
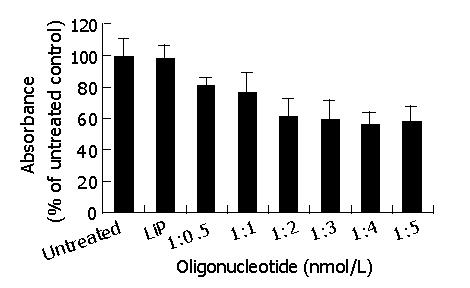
With the aim of obtaining the highest transfection efficiency and low non-specific effects, the compounds were generated using the antisense phosphorothioate oligonucleotides (ODNs) and highly effective Lipofectamine 2000 (LiP) by varying ODNs (μg):LiP (μL) ratios from 1:0.5 to 1:5. The ODNs designed by Olie et al[22], which targeted nucleotides (232-251) revealed the strongest effect. MTT was performed to test the effect of those compounds in surviving-HepG2 high-expression cells. As shown in Figure 1, the antisense compounds (1:4, 1:5) were identified as the most potent compounds. There was no significant difference in the effect between the compounds (1:4) and (1:5). Because the antisense compounds (1:0.5-1:3) could not achieve the best effect, and LiP had slight cytotoxicity, we only used the antisense compounds (1:4) in the following experiments.
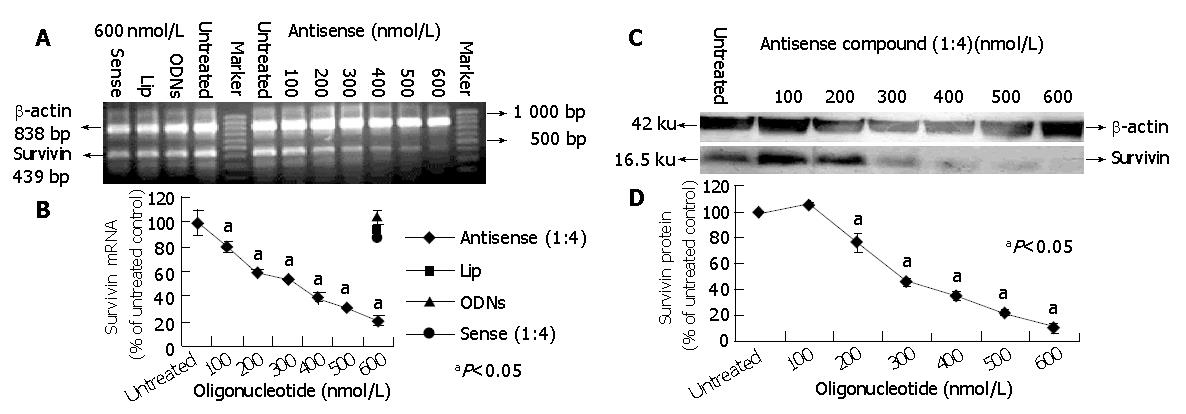
To further characterize the potency of antisense compounds (1:4) and their dose dependent effect on survivin expression levels (mRNA and protein) in HepG2 cells, the cells highly expressing survivin were transfected with different concentrations of antisense compounds (100-600 nmol/L), whereas the cells transfected with sense compound (600 nmol/L), LiP (600 nmol/L) and untreated cells were used as controls. Forty-eight hours after the start of transfection, the cells were examined by RT-PCR and Western blot. As shown in Figure 2, antisense compounds (1:4) down-regulated the survivin expression level in a dose-dependent manner with an IC50 of about 250 nmol/L. At a concentration of 500 nmol/L, a maximum down-regulation to 20% of the initial mRNA level was achieved. A further increase in oligonucleotide concentration did not result in increased antisense efficacy. The sense compound (600 nmol/L), LiP (600 nmol/L) and antisense compound controls without LiP (600 nmol/L) did not down-regulate the survivin expression level.
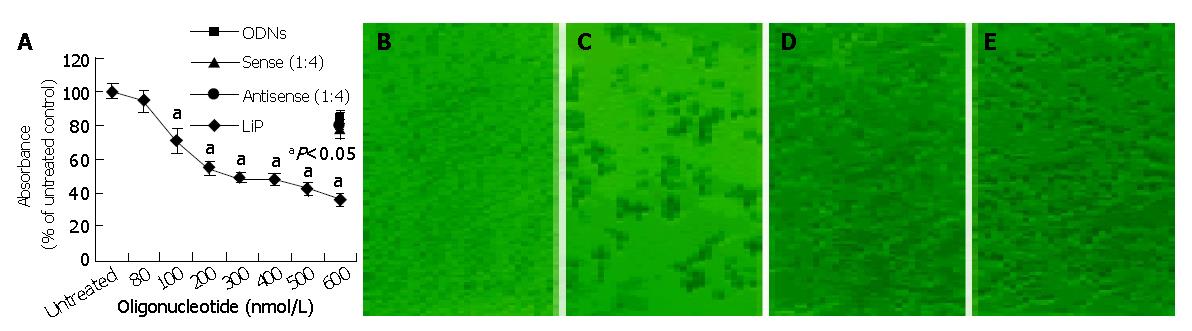
To analyze the biological effect associated with the down-regulation of survivin expression, the growth of HepG2 cells treated with antisense compounds was investigated by MTT assay. As shown in Figure 3A, 48 h after the start of transfection, antisense compounds reduced the growth of HepG2 cells dose-dependently, with an IC50 of 300 nmol/L. The unspecific growth-inhibitory effect of the sense compound (600 nmol/L) control was comparatively low, LiP (600 nmol/L) control had no growth-inhibitory effect. Antisense compounds induced death in HepG2 cells, as revealed by detachment from the culture surface (Figure 3: B, C, D, E).
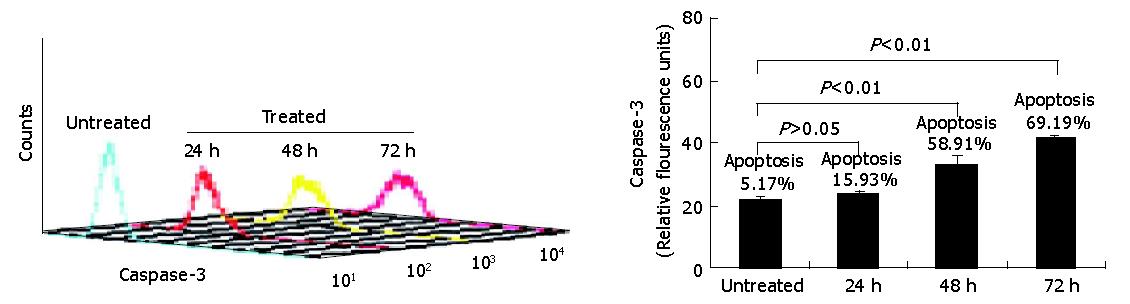
Having demonstrated that down-regulation of survivin expression reduced the viability of HepG2 cells, we analyzed whether cell death was due to the induction of apoptosis. As shown in Figure 4, HepG2 cells were treated with antisense compounds 24, 48, 72 h after the start of transfection. Then, the cells were analyzed by flow cytometry using PE-conjugated polyclonal rabbit anti-active caspase-3 antibody. Antisense compound-treated cells revealed increased caspase-3-like protease activity compared with untreated cells. Cells treated for 24 h had detectable active caspase-3 corresponding to a measurable value 23, whereas about 16% of the cells were induced to undergo apoptosis. Cells treated for 48 h had detectable active caspase-3 corresponding to a measurable value 33 corresponding to a 60% apoptosis ratio, while cells treated for 72 h had a measurable value 42 corresponding to a 70% apoptotic ratio. Untreated cells as control were primarily negative for the presence of active-caspase-3, a measurable value 16 corresponding to 5% apoptotic cells.
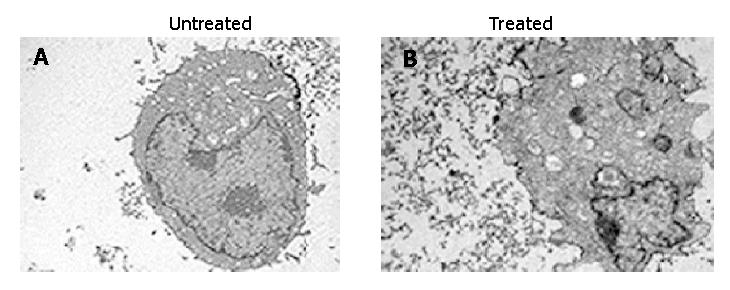
In order to confirm the cell apoptosis induced by treatment with antisense compounds, the morphological signs of apoptosis were evaluated by transmission electron microscopy. As shown in Figure 5, untreated HepG2 cells showed normal morphology in the nuclei and cytoplasm (Figure 5A). Treatment of HepG2 cells with antisense compounds resulted in changes such as blebbing and loss of microvilli, vacuolation in cytoplasm, condensation of cytoplasm and nuclei, and fragmented chromatin, providing further evidence for the induction of apoptosis as a consequence of survivin antisense treatment(Figure 5B).
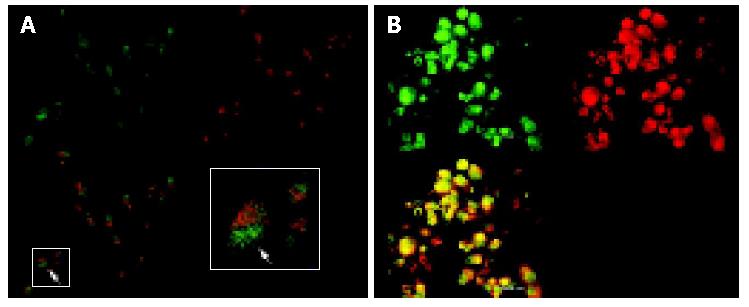
We carried out double immunofluorescence staining using FITC-labelled antibody against survivin and Alexa-labelled concanavalin A, with which cells of the membranes were counterstained. The stained cells were investigated by confocal laser scanning microscopy. As shown in Figures 6A,B in cells untreated with antisense compounds, green-immunofluorescence staining of survivin clearly showed that survivin was expressed mainly in the form of a spotted distribution inside the cytoplasm, which was named as survivin pool (Figure 6A). In cells treated with antisense compounds, however, there was rare and weak green fluorescence inside the cells, of which the membranes were counterstained with Alexa -labeled concanavalin A (Figure 6B).
Survivin, a member of the inhibitors of apoptosis protein family, deserves growing attention as “an ideal target” for cancer therapy[23,24] due to its differential expression in tumors vs normal tissues, distribution of subcellular localization, and the evidence for a dual role in both cell apoptosis and proliferation. It is expressed during embryonal development, lacks expression in terminally differentiated adult tissues, and becomes reexpressed in transformed cell lines and a variety of human tumors, with highest levels in liver cancer[4,20]. The expression of survivin in tumors is correlated with a shorter survival of patients with liver cancer, non-small cell lung cancer, colorectal cancer. Despite survivin represents an attractive target for therapy[25,26], the promise of survivin antisense ODNs to facilitate apoptosis of HepG2 cells, localization of survivin, and how the localization of survivin proteins is affected by transfection of antisense oligonucleotides in HepG2 cells remain to be determined. In the present study, we generated and optimized sequence-specific antisense compounds to down-regulate survivin expression (mRNA and protein) in HepG2 cell lines to demonstrate its ability to induce apoptosis, and how to localize survivin protein affected by antisense compounds.
Antisense approaches are efficient tools for the inhibition of gene expression in a sequence specific way[27]. Most antisense approaches rely on empirical targeting of oligonucleotides against the translation initiation site of mRNA, where the ATG start codon lies. The rationale for choosing this site is that it likely presents in single-stranded conformation, thus being accessible to antisense oligonucleotides. In a study by Olie et al[22], survivin antisense oligonucleotides, which target nucleotides 232-251, were identified, and most efficiently down-regulated the survivin mRNA level and directly induced apoptosis. In our study, we successfully transfected HepG2 cells with the above antisense oligonucleotides incorporating a vector of LiP, and generated and optimized the compounds by varying ODNs:LiP ratios from 1:0.5 to 1:5. Moreover, we identified that the compounds (1:4) were the most potent. As measured by RT-PCR and Western blot, the antisense compounds (1:4) also most efficiently down-regulated the survivin expression level in HepG2 cells, achieving its maximum effect at a concentration of 500 nmol/L, at which the expression level of survivin was down-regulated by 80%. By transfection of these compounds, we were also able to witness increased caspase-3-like protease activity by flow cytometric analysis, and morphological changes of apoptosis induced by antisense compounds such as vacuolation in cytoplasm, condensation of cytoplasm and nuclei and fragmented chromatin, with a parallel inhibition seen in cell proliferative activity by MTT assay.
A previous study reported that survivin could prevent apoptosis by targeting the terminal effectors caspase-3 and caspase-7, which act downstream in two major apoptotic pathways [3]. Survivin is believed to be expressed in the G2/M phase of the cell cycle in a cell cycle-regulated manner and is associated with microtubule formation of the mitotic spindle[10]. Antisense compounds (1:4) induce a strong growth-inhibitory effect and apoptosis in HepG2 cells in the absence of any further cytotoxic stimulus. These data suggest that targeting survivin with transfection of antisense ablates expression of endogenous survivin. However, loss of survivin expression is sufficient to trigger caspase-dependent apoptosis in HepG2 cells. Moreover, it has been suggested that the survivin antisense approach may be able to facilitate apoptosis through the two major apoptotic pathways. This observation is in agreement with the findings of others describing the necessity of interaction between survivin and microtubules of the mitotic spindle apparatus to prevent a default induction of apoptosis at the G2/M phase of cell cycle[10].
One of the most significant features of survivin is expressed in the G2/M phase of cell cycle in a cell-regulated manner. At the beginning of mitosis, survivin is associated with microtubules of the mitotic spindle in a specific and saturable reaction that is regulated by microtubule dynamics[10]. Recent studies have suggested an alternative distribution of survivin localized to kinetochores, centrosomes (microtubule-organizing centers), spindle microtubules, central spindle midzone and midbodies. Approximately 80% of the total cellular survivin content in mitotic cells is bound to centrosomes and microtubules of the metaphase and anaphase spindles. Survivin’s localizations in the mitotic spindle apparatus might be essential for anti-apoptotic function[28].
In the present study, double immunofluorescence analysis has confirmed the presence of survivin protein pool in cytoplasm. In untreated HepG2 cells, labeled-FITC-immunofluorescence staining of survivin clearly showed that survivin was expressed mainly in the form of a spotted distribution inside the cytoplasm. Whereas, in treated cells, survivin was rare and weak in immunoreactivity. As demonstrated by Western blot, in cells treated with antisense compounds, survivin expression level was in a dose-dependent manner. It is suggested that the survivin antisense approach may be able to affect the localization of survivin. Recent experiments demonstrate that survivin exists in an immunochemically distinct pool of survivin[10]. A more recent study suggests that overexpression of survivinDsRed fusion proteins is diffusely distributed in cytoplasm with distinct spots[29]. We also found that survivin was expressed in cytoplasm in the form of pools or spots, suggesting that survivin plays an important role in linking cell death and proliferation. In the context, antisense disruption of survivin-microtubule interactions can result in two effects during mitosis: increased caspase-3 activity-a mechanism involved in cell death and loss of anti-apoptosis function of survivin.
In summary, down-regulation of survivin expression by antisense compounds facilitates apoptosis, and localization of survivin proteins is affected by antisense compounds in HepG2 cells. Survivin protein is a key molecule connecting cell proliferation with apoptosis. Antisense targeting survivin has a bright prospect in liver cancer therapy.
| 1. | Makin G, Hickman JA. Apoptosis and cancer chemotherapy. Cell Tissue Res. 2000;301:143-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 162] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 2. | Adida C, Crotty PL, McGrath J, Berrebi D, Diebold J, Altieri DC. Developmentally regulated expression of the novel cancer anti-apoptosis gene survivin in human and mouse differentiation. Am J Pathol. 1998;152:43-49. [PubMed] |
| 3. | Tamm I, Wang Y, Sausville E, Scudiero DA, Vigna N, Oltersdorf T, Reed JC. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998;58:5315-5320. [PubMed] |
| 4. | Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917-921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2342] [Cited by in RCA: 2401] [Article Influence: 82.8] [Reference Citation Analysis (1)] |
| 5. | Deveraux QL, Reed JC. IAP family proteins--suppressors of apoptosis. Genes Dev. 1999;13:239-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1910] [Cited by in RCA: 1910] [Article Influence: 70.7] [Reference Citation Analysis (0)] |
| 6. | LaCasse EC, Baird S, Korneluk RG, MacKenzie AE. The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene. 1998;17:3247-3259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 728] [Cited by in RCA: 716] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 7. | Verdecia MA, Huang H, Dutil E, Kaiser DA, Hunter T, Noel JP. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat Struct Biol. 2000;7:602-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 566] [Cited by in RCA: 596] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 8. | Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2467] [Cited by in RCA: 2477] [Article Influence: 95.3] [Reference Citation Analysis (4)] |
| 9. | Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1742] [Cited by in RCA: 1697] [Article Influence: 65.3] [Reference Citation Analysis (0)] |
| 10. | Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1396] [Cited by in RCA: 1453] [Article Influence: 51.9] [Reference Citation Analysis (0)] |
| 11. | Skoufias DA, Mollinari C, Lacroix FB, Margolis RL. Human survivin is a kinetochore-associated passenger protein. J Cell Biol. 2000;151:1575-1582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 188] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 12. | Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL, Choo KH. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol. 2000;10:1319-1328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 402] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 13. | Wheatley SP, Carvalho A, Vagnarelli P, Earnshaw WC. INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr Biol. 2001;11:886-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 212] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 14. | Lu CD, Altieri DC, Tanigawa N. Expression of a novel antiapoptosis gene, survivin, correlated with tumor cell apoptosis and p53 accumulation in gastric carcinomas. Cancer Res. 1998;58:1808-1812. [PubMed] |
| 15. | Zhu XD, Lin GJ, Qian LP, Chen ZQ. Expression of survivin in human gastric carcinoma and gastric carcinoma model of rats. World J Gastroenterol. 2003;9:1435-1438. [PubMed] |
| 16. | Monzó M, Rosell R, Felip E, Astudillo J, Sánchez JJ, Maestre J, Martín C, Font A, Barnadas A, Abad A. A novel anti-apoptosis gene: Re-expression of survivin messenger RNA as a prognosis marker in non-small-cell lung cancers. J Clin Oncol. 1999;17:2100-2104. [PubMed] |
| 17. | Tanaka K, Iwamoto S, Gon G, Nohara T, Iwamoto M, Tanigawa N. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer Res. 2000;6:127-134. [PubMed] |
| 18. | Kato J, Kuwabara Y, Mitani M, Shinoda N, Sato A, Toyama T, Mitsui A, Nishiwaki T, Moriyama S, Kudo J. Expression of survivin in esophageal cancer: correlation with the prognosis and response to chemotherapy. Int J Cancer. 2001;95:92-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 19. | Yoshida H, Ishiko O, Sumi T, Matsumoto Y, Ogita S. Survivin, bcl-2 and matrix metalloproteinase-2 enhance progression of clear cell- and serous-type ovarian carcinomas. Int J Oncol. 2001;19:537-542. [PubMed] |
| 20. | Ito T, Shiraki K, Sugimoto K, Yamanaka T, Fujikawa K, Ito M, Takase K, Moriyama M, Kawano H, Hayashida M. Survivin promotes cell proliferation in human hepatocellular carcinoma. Hepatology. 2000;31:1080-1085. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 266] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 21. | Grossman D, McNiff JM, Li F, Altieri DC. Expression of the apoptosis inhibitor, survivin, in nonmelanoma skin cancer and gene targeting in a keratinocyte cell line. Lab Invest. 1999;79:1121-1126. [PubMed] |
| 22. | Olie RA, Simões-Wüst AP, Baumann B, Leech SH, Fabbro D, Stahel RA, Zangemeister-Wittke U. A novel antisense oligonucleotide targeting survivin expression induces apoptosis and sensitizes lung cancer cells to chemotherapy. Cancer Res. 2000;60:2805-2809. [PubMed] |
| 23. | Jäättelä M. Escaping cell death: survival proteins in cancer. Exp Cell Res. 1999;248:30-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 464] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 24. | Kawasaki H, Altieri DC, Lu CD, Toyoda M, Tenjo T, Tanigawa N. Inhibition of apoptosis by survivin predicts shorter survival rates in colorectal cancer. Cancer Res. 1998;58:5071-5074. [PubMed] |
| 25. | Nakagawara A. Molecular basis of spontaneous regression of neuroblastoma: role of neurotrophic signals and genetic abnormalities. Hum Cell. 1998;11:115-124. [PubMed] |
| 26. | Saitoh Y, Yaginuma Y, Ishikawa M. Analysis of Bcl-2, Bax and Survivin genes in uterine cancer. Int J Oncol. 1999;15:137-141. [PubMed] |
| 27. | Urban E, Noe CR. Structural modifications of antisense oligonucleotides. Farmaco. 2003;58:243-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 66] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 28. | Shin S, Sung BJ, Cho YS, Kim HJ, Ha NC, Hwang JI, Chung CW, Jung YK, Oh BH. An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and -7. Biochemistry. 2001;40:1117-1123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 510] [Cited by in RCA: 532] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 29. | Temme A, Rieger M, Reber F, Lindemann D, Weigle B, Diestelkoetter-Bachert P, Ehninger G, Tatsuka M, Terada Y, Rieber EP. Localization, dynamics, and function of survivin revealed by expression of functional survivinDsRed fusion proteins in the living cell. Mol Biol Cell. 2003;14:78-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
Edited by Wang XL and Zhu LH