Published online May 7, 2005. doi: 10.3748/wjg.v11.i17.2597
Revised: September 14, 2004
Accepted: December 3, 2004
Published online: May 7, 2005
AIM: Interferon α2b (IFNα2b) and thymosin α1 (Tα1) exhibit synergic effects in the treatment of hepatitis B and hepatitis C when used together. For developing a fusion protein drug, fusion proteins of IFNα2b and Tα1 linked by different lengths of (G4S)n (n = 1-3) were constructed and expressed in Pichia pastoris.
METHODS: Using PCR and molecular clone techniques, the fusion genes of IFNα2b-(G4S)n-Tα1 (n = 1-3) were constructed and subcloned into the eukaryotic expression vector pPIC9. After transformation of these plasmids into P. pastoris, the expressed fusion proteins IFNα2b-(G4S)n-Tα1 (n = 1-3) were obtained. These proteins were purified through diethylaminoethyl (DEAE) affinity chromatography and Superdex™ 75 gel filtration and analyzed by SDS-PAGE and Western blot. Antiviral and E-rosette assays were used to investigate the bioactivities of these fusion proteins.
RESULTS: DNA sequencing confirmed that the fusion genes of IFNα2b-(G4S)n-Tα1 (n = 1-3) were correctly cloned to the pPIC9 vector. The recombinant IFNα2b-(G4S)n-Tα1 (n = 1-3) fusion proteins expressed in P. pastoris were purified with DEAE and Superdex™ 75 gel filtration chromatography. The fusion proteins could be observed on sodium dodecylsulfate-polyacrylamide gel electrophoresis with molecular weight (MW) of 23.2, 22.9, and 22.6 ku, respectively, and reacted to the IFNα2b monoclonal antibody and Tα1 polyclonal antibody. The purified fusion proteins exhibit antiviral activity and can enhance the percentage of E-rosette-forming-cell in E-rosette assay.
CONCLUSION: The recombinant IFNα2b-(G4S)n-Tα1 (n = 1-3) fusion proteins were successfully expressed in P. pastoris. Purified fusion proteins exhibit both antiviral activity of IFNα2b and immunomodulatory activity of Tα1 in vitro. These results will be the basis for further evaluation of the fusion proteins’ function in vivo.
- Citation: Yang YF, Yuan HY, Liu NS, Chen XL, Gao BY, Lu H, Li YY. Construction, expression and characterization of human interferon α2b-(G4S)n-thymosin α1 fusion proteins in Pichia pastoris. World J Gastroenterol 2005; 11(17): 2597-2602
- URL: https://www.wjgnet.com/1007-9327/full/v11/i17/2597.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i17.2597
The human interferon α2b (IFNα2b), the most widely used member of IFNα family, exerts many biological actions including broad-spectrum antiviral effects, inhibition of tumor cell proliferation and enhancement of immune functions[1-3]. The human thymosin α1 (Tα1) plays an important role in the maturation, differentiation and function of lymphocytes[4,5]. Both are widely used proteins in the treatment of hepatitis B and C[5-8]. A series of clinical trails show that, when compared with standard therapeutic regimens, combination therapy of IFNα and Tα1 could significantly enhance virological and biochemical response rates in patients with hepatitis B and C, and is well tolerated[9-12]. Combination of Tα1 with IFNα is becoming one of the most promising options in improving the response rate of chronic hepatitis B virus and hepatitis C virus infection and decreasing its probability of developing into hepatocellular carcinoma[13]. To decrease the high cost of current combination therapy, we tried to express the IFNα2b and Tα1 fusion protein to use as a substitute.
The human recombinant IFNα2b has been successfully expressed and used in clinics for more than 10 years[14]. The crystal structure of IFNα2b has been determined, and many mutation researches have been reported[15,16]. However, no recombinant Tα1 has been expressed or applied into clinical use successfully. The Tα1 currently used in therapy of hepatitis is chemically synthesized[5]. The structure of Tα1 has been studied using circular dichroism and two-dimensional NMR[17]. This information provides guidance in designing of the IFNα2b and Tα1 fusion proteins. To keep the independence of protein folding and function, the flexible G4S immunosilent amino acid sequence was adopted to link the C terminal of IFNα2b and the N terminal of Tα1, a sequence widely used in domain ligation[18,19]. The molecular design of NH3-IFNα2b-(G4S)n-Tα1-COOH (n = 1-3) was set down. The powerful Pichia pastoris/pPIC9 eukaryotic expression system was employed to express the fusion proteins, which could grow rapidly at high densities and carries the strong alcohol oxidase (AOX1) I promoter, the mating factor signal sequence from S. cerevisiae and the His4 selectable marker (Invitrogen, 131020sa).
In the present study, IFNα2b-(G4S)n-Tα1 (n = 1-3) fusion genes were constructed and expressed in P. pastoris. Their products were isolated by affinity chromatography and gel filtration. Primary bioactivity assay of fusion proteins is described and compared with standard samples of IFNα2b and Tα1, respectively.
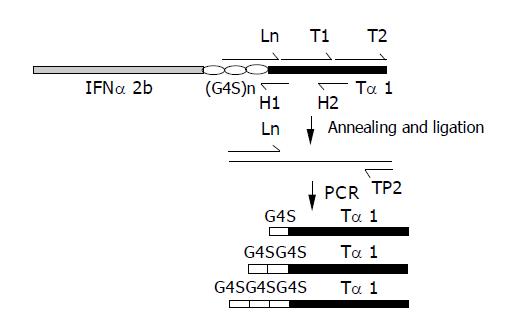
The coding strand of (G4S)3-Tα1 was synthesized as three successive single-strand DNA fragments: L1, T1 and T2 (Bioasia Biotechnologies, Inc., Shanghai). To ligate the three fragments together, two complementary short non-coding strands of H1 and H2 were synthesized, which are complementary to the L1-T1 and T1-T2 fusion joints separately (Figure 1). After phosphorylation of T1 and T2, equal amounts of L1, T1, T2, H1 and H2 were mixed and incubated at 100 °C for 2 min, then annealed at room temperature. The annealed fragments were ligased overnight at L1-T1 and T1-T2 joints using T4-DNA ligase (New England Biolabs) at 14 °C. This reaction mixture was then precipitated in 100% ethanol, dried and re-suspended in water. Using the re-suspended ligation product as template, L1 (5’- GGTGGCGGATCCGGCGGTGGTGGTTCT-GGTGGCGGCGGCTCTTCATC-3’) and TP2 (5’-GGAAGCTTAATTTTCTGCCTCTTC-3’) as primers, which introduced a BamHI site at the 5’-end and an HindIII site at the 3’-end of the fragments, (G4S)3-Tα1 was amplified in a standard PCR protocol. This reaction was performed using a Perkin-Elmer DNA thermal cycler. The experimental conditions included 1 cycle at 95 °C for 5 min, 25 cycles at 94 °C for 30 s, 58 °C for 30 s, 72 °C for 2 min, followed by 72 °C for 10 min. The PCR product was purified by electrophoresis on 8 g/L agarose with a gel extraction kit (Watson Biotechnologies, Inc., Shanghai) and digested with BamHI and HindIII. The digested fragment (G4S)3-Tα1 was purified and ready for further ligation. Fragments (G4S)2-Tα1 and G4S-Tα1 were constructed in the same way and were also digested with BamHI and HindIII and purified by means of a gel extraction kit.
The Ln represents L1 (5’-GGTGGCGGATCCG-GCGGTGGTGGTTCTGGTGGCGGCGGCTCTTCATC-3’), L2 (5’-GGTGGCGGATCCGGCGGCGGTGGT-TCATC-3’) and L3 (5’-GGTGGCGGATCCTCATC-3’), introduced of a BamHI cleavage site.
Ln, H1 (5’-AACGGCTGCGTCTGATGAACCACC-3’), H2 (5’-CTTCTTCTCCTTTAAGTCC-3’) and phosphorylated T1 (5’-AGACGCAGCCGTTGACACTAGCTCTGAAA-TCACTACTAAGGACTTAAAG-3’) and T2 (5’-GAGAAG-AAGGAAGTTGTGGAAGAGGCAGAAAATTAAGCTTCC-3’) were annealed. While the Ln, T1 and T2 were ligased to use as the template. In the guide of Ln and TP2 (5’-GGAAGCTTAATTTTCTGCCTCTTC-3’), which has a HindIII cleavage site, the (G4S)n-thymosin α1 (n = 1-3) fragments were amplified.
The coding sequence of the IFNα2b gene was amplified by PCR using IFNα2b-containing plasmid CB104 as template with IP1 (5’-GGGAATTCTGTGATCTGCCT-CAAAC-3’) and IP2 (5’-CCGGATCCGCCACCGCCTTC-CTTACTTCTTAAACTTTCTTGC-3’) as primers, in which EcoRI and BamHI were introduced to 5’ and 3’ end, respectively. The PCR was performed by employing the following temperature cycle: 1 cycle at 95 °C for 5 min, 25 cycles at 94 °C for 30 s, 58 °C for 45 s, 72 °C for 2 min, followed by 72 °C for 10 min. The amplified PCR product was purified by electrophoresis on 8 g/L agarose with a gel extraction kit (Watson Biotechnologies, Inc., Shanghai). The PCR product was digested with EcoRI and BamHI and purified for ligation reaction.
Both IFNα2b and (G4S)3-Tα1 DNA digested fragments were inserted together into the HindIII and EcoRI digested pUC118 vector. The ligation products were transformed into the TOP10 E. coli strain and selected on amplicillin-containing LB plates. The recombinant plasmid was then extracted from transformant and confirmed by nucleotide sequencing (377 DNA sequencer, Bioasia Biotechnologies, Inc., Shanghai), finally obtaining the plasmid pUC118-IFNα2b-(G4S)3-Tα1 with correct reading frame. Using the same strategy, the two recombinant plasmids, pUC118-IFNα2b-(G4S)2-Tα1 and pUC118-IFNα2b-G4S-Tα1, were also constructed.
The encoding sequence of IFNα2b-(G4S)3-Tα1 was amplified using EP1 (5’-TCTCTCGAGAAAAGATG-TGATCTGCCTCAAAC-3’) and EP2 (5’-CGCGGCCG-TTAATTTTCTGCCTCTTC-3’) as primers, digested by XhoI and NotI and inserted into the P. pastoris expression vector pPIC9. The resultant plasmid pPIC9-IFNα2b-(G4S)3-Tα1 was confirmed by the nucleotide sequencing. The plasmids pPIC9-IFNα2b-(G4S)2-Tα1 and pPIC9-IFNα2b-G4S-Tα1 were obtained in the same way.
The P. pastoris strain SMD1168 (his4, pep4) (Invitrogen, 131020sa) was employed to express the fusion proteins. The pPIC9-IFNα2b-(G4S)n-Tα1 (n = 1-3) plasmids were linearized by SalI and transformed into P. pastoris (SMD1168) via electroporation (Eppendorf, electroporator 2510). The transformants were selected according to the manufacturer’s direction (Invitrogen, 131020sa). The obtained engineering strains were named SMD1168/pPIC9-IFNα2b-(G4S) 3-Tα1, SMD1168/pPIC9-IFNα2b-(G4S)2-Tα1, and SMD1168/pPIC9-IFNα2b-G4S-Tα1, respectively. Each engineering strain was incubated in the standard conditions as described by the manufacturer (Invitrogen, 131020sa). After induction of methanol, the supernatant of the culture was collected with 5 min of 10000 g and monitored by 15% SDS-PAGE. Proteins were visualized by 2.5 g/L Coomassie brilliant blue R-250 staining. The proportion of the expressed fusion protein in all the secreted proteins was analyzed by computer UVR ware (Shanghai Tanon Science and Technology Co. Ltd). Another gel of SDS-PAGE was transblotted onto a PVDF membrane (Millipore Corporation) for Western blotting. The fusion proteins were then detected by incubating the transblotted PVDF membrane with an anti-IFNα2b monoclonal antibody followed by a HRP-conjugated goat anti-mouse antibody (PIERCE) and anti-Tα1 polyclonal antibody (Sigma, St. Louis, MO) followed by a HRP-conjugated goat anti-rabbit antibody (PIERCE), respectively. The immunoreactive protein was visualized by DAB (Roche).
The collected supernatant liquid of 0.05-L culture was dialyzed at 4 °C overnight against the dialysis buffer (0.05 mol/L Tris-HCl, 0.01 mol/L NaCl, pH 8.0), and then applied to a DEAE column (2.5 cm×12 cm) equilibrated with the equilibration buffer (0.05 mol/L Tris-HCl, 0.01 mol/L NaCl, pH 9.0). The fusion protein was eluted sequentially with the elution buffer A (0.05 mol/L Tris-HCl, 0.10 mol/L NaCl, pH 8.0), and elution buffer B (0.05 mol/L Tris-HCl, 0.20 mol/L NaCl, pH 8.0). The elution fractions containing expressed fusion protein were pooled and applied to Superdex™ 75 (Amersham Pharmacia Biotech, Uppsala, Sweden) columns (2.5 cm×15 cm) for the further purification. The chromatography was performed at 0.5×10-3 L/fraction at room temperature. The protein was monitored by measuring the UV absorbency at 280 nm (Amersham Pharmacia Biotech, Uppsala, Sweden). And the pooled elution fractions containing the fusion protein from Superdex™ 75 column were analyzed by SDS-PAGE. Density quantification analysis was used to quantify the proportion of purified proteins among the eluates. Finally, the purified fusion protein was collected and stored at -20 °C for detection of bioactivities.
The human amnion cells (WISH) were seeded into 96-well plates (3×104 cells/well) and incubated overnight at 37 °C, 50 mL/L CO2 in a humidified incubator. Then dilutions of standard samples of IFNα2b (Wanxing Pharmical Tech. Ltd, Shanghai, China) and the purified fusion proteins IFNα2b-(G4S)n-Tα1 (n = 1-3), all having the same concentration, were added. The cells were then incubated for 24 h. Following this vesicular stomatitis virus (VSV) was added, 1.2×105 plaque forming units per well, and the cells allowed continued incubation for another 40 h. Ten microliters of 5 g/L MTT solution (Sigma Cat. No. M5655) was then added into each well. After incubation for another 4 h at 37 °C, 50 µL of MTT lysing solution (20% SDS 50% DMF) was added to each well and incubated overnight. The plates were read at 570 nm and the unit of each sample’s antiviral activity was calculated.
Fresh pig thymus was cut and rubbed. Cells were then separated by lymphocyte separation medium (Shanghai Reagent Factory) and placed with HRPMI-1640 and fusion proteins for 2 h at 37 °C, 50 mL/L CO2 in a humidified incubator, using Tα1 (SciClone Pharmaceuticals, Inc., CA, USA), IFNα2b and cells with no drug added as parallel controls. Sheep red blood cells were then added and incubation continued for another 3 h. The percentage of E-rosette forming lymphocytes was calculated by counting the number of E-rosette forming lymphocytes out of a total of 200 lymphocytes. The experiment was repeated five times with a sample concentration of 2.6×10-17 mol/L.
As described in Materials and Methods, pPIC9-IFNα2b-G4S-Tα1, pPIC9-IFNα2b-(G4S)2-Tα1 and pPIC9-IFNα2b-(G4S)3-Tα1 were constructed and identified by the PCR method using EP1 and EP2 as primers, respectively. The bands specific to IFNα2b-G4S-Tα1, IFNα2b-(G4S)2-Tα1 and IFNα2b-(G4S)3-Tα1 were 614, 629, and 644 bp respectively (Figure 2). Nucleotide sequencing confirmed that the three constructs were all in the correct reading frame.
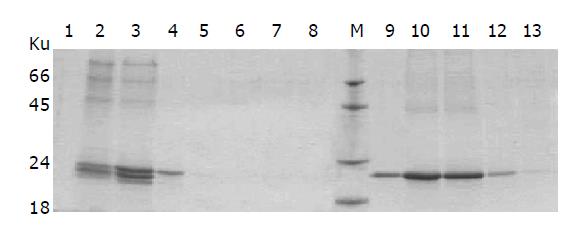
The linearized pPIC9-IFNα2b-(G4S)n-Tα1 (n = 1-3) plasmids were transformed separately into P. pastoris (SMD1168) and the transformants of pPIC9-IFNα2b-(G4S)-Tα1, pPIC9-IFNα2b-(G4S)2-Tα1 and pPIC9-IFNα2b-(G4S)3-Tα1 were incubated separately in the standard fermentation medium (Invitrogen, 131020sa) and induced by methanol. After 48 h of induction, the supernatants of the culture were collected by centrifugation and analyzed by SDS-PAGE. As shown in Figure 3A, the bands at 23.2, 22.9, and 22.6 ku, produced separately by the three engineering strains, are specific compared with control SMD1168/pPIC9. Density quantification analysis shows that the fusion proteins comprise 37.24% of the total secreted protein (Figure 3B), approximately 0.22 g/L per 0.03 L of shake culture. Using a monoclonal antibody directly towards IFNα2b and a polyclonal antibody towards Tα1, Western blot analysis confirms the expression of the IFNα2b-(G4S)n-Tα1 fusion proteins (Figure 4). Both SDS-PAGE and Western blot analysis show two expressed protein bands on each lane which are not found on control: a main protein band and an additional protein band. The main protein bands just show approximately the calculated MW: 23.2 ku for IFNα2b-(G4S)3-Tα1, 22.9 ku for IFNα2b-(G4S)2-Tα1 and 22.6 ku for IFNα2b-G4S-Tα1, and the additional protein bands, with a lower MW, migrate at a slightly faster rate than the major protein bands. When the supernatant was left at room temperature for a long time, the supernatant’s low-molecular-weight proteins increase. So we believe, preliminarily, that the low-molecular-weight band is a degradation fragment of the fusion protein. Although the P. pastoris strain SMD 1168 is pep4 protease deficient, there may be other mechanisms involved in this process. Subsequent research will be done to find the protein degradation mechanism and methods to stabilize their fusions. No visible higher MW fusion proteins are observed. This sometimes happens in yeast expression, suggesting glycosylation modification.
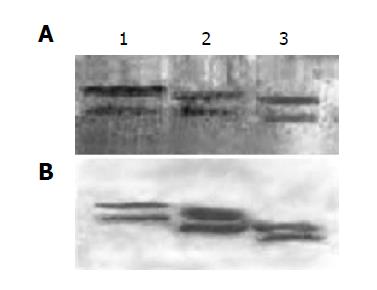
DEAE affinity chromatography was employed to purify fusion proteins of IFNα2b-(G4S)n-Tα1 (n = 1-3). After dialysis against the dialysis buffer, the supernatant of the culture was applied to the DEAE column, which was then eluted by the elution buffer A followed by the elution buffer B. Nearly all of the degraded fragment of fusion protein and part of the integrated fusion protein were eluted from DEAE column by elution buffer A (0.05 mol/L Tris-HCl, 0.10 mol/L NaCl, pH 8.0). When the concentration of NaCl was enhanced in elution buffer B (0.05 mol/L Tris-HCl, 0.20 mol/L NaCl, pH 8.0), most of the integrated fusion protein was eluted (Figure 5); this indicates that at pH 8.0 the integrated protein binds the DEAE column tighter than the degradation fragment. The eluted fractions containing the integrated fusion protein were then pooled together and applied to Superdex™ 75 gel filtration for further purification. The purified IFNα2b-(G4S)n-Tα1 (n = 1-3) fusion proteins were found to migrate as single bands in Coomassie blue-stained SDS-PAGE (Figure 6A). Density quantification analysis showed that the purified fusion protein is a single peak with purity of more than 98% (Figure 6B).
The antiviral activity of the purified IFNα2b-(G4S)n-Tα1 (n = 1-3) proteins was measured using the standard cytopathic protection assay. IFNα2b could be used to protect the cultured WISH cells from VSV destruction. In this experiment, IFNα2b-G4S-Tα1, IFNα2b-(G4S)2-Tα1, and IFNα2b-(G4S)3-Tα1 exhibit antiviral activities of about 3.17×107, 1.75×107, and 3.22×107 U/mg, respectively. The antiviral activity of IFNα2b-(G4S)3-Tα1 is the highest.
Tα1 is active in enhancing the production of receptors for sheep erythrocytes by T-cells[20,21]. So Tα1 standard sample could enhance the percentage of E-rosette-forming-cell (ERFC) in E-rosette assay. IFNα2b-(G4S)n-Tα1 (n = 1-3) also exhibits such activity. As shown in Figure 7, ERFC of the fusion proteins is slightly lower than that of Tα1 standard sample, but is higher than that of IFNα2b. According to the references, IFNα2b has negative or slightly positive effects on the formation of ERFC[22,23]. So it reflects that Tα1 could exhibit some of its function in the fusion protein. IFNα2b-(G4S)2-Tα1 produced ERFC near the same rate as Tα1, higher than that of other fusion proteins.
Construction of fusion protein is widely used in cytokine research. Many fusion proteins such as recombinant human albumin-IFNα2b, interferon γ-tumor necrosis factor β, interferon-γ-gp120, etc. have been expressed and some of them have been used in clinical trails[24-27]. Within the same tradition, we expressed here one set of IFNα2b and Tα1 fusion proteins.
The biological properties of the IFNα2b-(G4S)n-Tα1 fusion proteins were studied using a standard antiviral and an E-rosette assay. These proteins exhibit antiviral activities ranging from 1.75×107 to 3.22×107 U/mg, lower than that of the standard IFNα2b (1.25×108 U/mg), though near to that of IFNγ (2.0×107 U/mg)[28]. The IFNα2b-(G4S)2-Tα1 fusion protein is comparable to Tα1 in its ability to produce ERFC. Presumably, IFNα2b-(G4S)2-Tα1 might best conserve the function of IFNα2b and Tα1, which would be emphasized in future experiment. Our results show that, among the different lengths of G4S-linked fusion proteins, their relationship is nonlinear in both antiviral activity and E-rosette forming promotion ability. This may partly be explained by the complication of protein folding.
In summary, fusion proteins of IFNα2b-(G4S)n-Tα1 (n = 1-3) can be expressed in P. pastoris and purified through DEAE column chromatography and Superdex™ 75 gel filtration with 98% purity. They are able to keep the antiviral activity of IFNα2b and similar to Tα1 can enhance the percentage of ERFC in E-rosette assay. Among these IFNα2b-(G4S)2-Tα1 was most effective. This work provides an excellent basis for further research on one kind of inexpensive hepatitis therapy drug.
We would like to thank Yi Zhang who is in Wanxing Pharmical Tech. Ltd for the determination of antiviral activities and Dr. William K. for reading the proofs.
| 1. | Di Bisceglie AM, Martin P, Lisker-Melman M, Kassianides C, Korenman J, Bergasa NV, Baker B, Hoofnagle JH. Therapy of chronic delta hepatitis with interferon alfa-2b. J Hepatol. 1990;11 Suppl 1:S151-S154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 2. | Tamayo L, Ortiz DM, Orozco-Covarrubias L, Durán-McKinster C, Mora MA, Avila E, Teixeira F, Ruiz-Maldonado R. Therapeutic efficacy of interferon alfa-2b in infants with life-threatening giant hemangiomas. Arch Dermatol. 1997;133:1567-1571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 28] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 3. | Romerio F, Zella D. MEK and ERK inhibitors enhance the anti-proliferative effect of interferon-alpha2b. FASEB J. 2002;16:1680-1682. [PubMed] |
| 4. | Ancell CD, Phipps J, Young L. Thymosin alpha-1. Am J Health Syst Pharm. 2001;58:879-85; quiz 886-8. [PubMed] |
| 5. | Billich A. Thymosin alpha1. SciClone Pharmaceuticals. Curr Opin Investig Drugs. 2002;3:698-707. [PubMed] |
| 6. | Rothstein KD, Munoz SJ. Interferon and other therapies for hepatitis B and hepatitis C infections. Clin Lab Med. 1996;16:465-491. [PubMed] |
| 7. | Molloy PJ, Azzouz M, Van Thiel DH. Treatment of chronic hepatitis B and hepatitis C with interferon alfa-2b. Am Fam Physician. 1996;54:1598-1605. [PubMed] |
| 8. | Arase Y, Tsubota A, Suzuki Y, Suzuki F, Kobayashi M, Someya T, Akuta N, Hosaka T, Saitoh S, Ikeda K. A pilot study of thymosin alpha1 therapy for chronic hepatitis B patients. Intern Med. 2003;42:941-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 9. | Saruc M, Yuceyar H, Kucukmetin N, Demir MA, Kandiloglu AR. Combination thymosin-alpha 1 and interferon-alpha 2b in the treatment of anti-HBe-positive chronic hepatitis B in Turkey. Hepatogastroenterology. 2002;49:798-802. [PubMed] |
| 10. | Saruc M, Ozden N, Turkel N, Ayhan S, Hock LM, Tuzcuoglu I, Yuceyar H. Long-term outcomes of thymosin-alpha 1 and interferon alpha-2b combination therapy in patients with hepatitis B e antigen (HBeAg) negative chronic hepatitis B. J Pharm Sci. 2003;92:1386-1395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 11. | Sherman KE, Sjogren M, Creager RL, Damiano MA, Freeman S, Lewey S, Davis D, Root S, Weber FL, Ishak KG. Combination therapy with thymosin alpha1 and interferon for the treatment of chronic hepatitis C infection: a randomized, placebo-controlled double-blind trial. Hepatology. 1998;27:1128-1135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 56] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 12. | Moscarella S, Buzzelli G, Romanelli RG, Monti M, Giannini C, Careccia G, Marrocchi EM, Zignego AL. Interferon and thymosin combination therapy in naive patients with chronic hepatitis C: preliminary results. Liver. 1998;18:366-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 13. | Rasi G, Pierimarchi P, Sinibaldi Vallebona P, Colella F, Garaci E. Combination therapy in the treatment of chronic viral hepatitis and prevention of hepatocellular carcinoma. Int Immunopharmacol. 2003;3:1169-1176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Baron E, Narula S. From cloning to a commercial realization: human alpha interferon. Crit Rev Biotechnol. 1990;10:179-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 15. | Radhakrishnan R, Walter LJ, Hruza A, Reichert P, Trotta PP, Nagabhushan TL, Walter MR. Zinc mediated dimer of human interferon-alpha 2b revealed by X-ray crystallography. Structure. 1996;4:1453-1463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 175] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 16. | Levy WP, Rubinstein M, Shively J, Del Valle U, Lai CY, Moschera J, Brink L, Gerber L, Stein S, Pestka S. Amino acid sequence of a human leukocyte interferon. Proc Natl Acad Sci USA. 1981;78:6186-6190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 93] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 17. | Grottesi A, Sette M, Palamara T, Rotilio G, Garaci E, Paci M. The conformation of peptide thymosin alpha 1 in solution and in a membrane-like environment by circular dichroism and NMR spectroscopy. A possible model for its interaction with the lymphocyte membrane. Peptides. 1998;19:1731-1738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Frankel AE, McCubrey JA, Miller MS, Delatte S, Ramage J, Kiser M, Kucera GL, Alexander RL, Beran M, Tagge EP. Diphtheria toxin fused to human interleukin-3 is toxic to blasts from patients with myeloid leukemias. Leukemia. 2000;14:576-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 87] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 19. | Thompson J, Stavrou S, Weetall M, Hexham JM, Digan ME, Wang Z, Woo JH, Yu Y, Mathias A, Liu YY. Improved binding of a bivalent single-chain immunotoxin results in increased efficacy for in vivo T-cell depletion. Protein Eng. 2001;14:1035-1041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 46] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Low TL, Thurman GB, McAdoo M, McClure J, Rossio JL, Naylor PH, Goldstein AL. The chemistry and biology of thymosin. I. Isolation, characterization, and biological activities of thymosin alpha1 and polypeptide beta1 from calf thymus. J Biol Chem. 1979;254:981-986. [PubMed] |
| 21. | Ahmed A, Wong DM, Thurman GB, Low TL, Goldstein AL, Sharkis SJ, Goldschneider I. T-lymphocyte maturation: cell surface markers and immune function induced by T-lymphocyte cell-free products and thymosin polypeptides. Ann N Y Acad Sci. 1979;332:81-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 63] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 22. | Avdeeva ZhI, Mazina NM, Krylov OR, Olisova MO, Lukasheva TV, Medunitsyn NV. The immunomodulating action of interferon. The change in the rosette-forming activity of human blood lymphocytes under the influence of gamma-interferon and recombinant alpha 2-interferon. Vestn Dermatol Venerol. 1990;4:16-19. [PubMed] |
| 23. | Fuhlbrigge RC, Edwards BS, Borden EC. Effects of interferon alpha on the sheep erythrocyte "receptor" of human lymphocytes. J Interferon Res. 1984;4:499-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 24. | Osborn BL, Olsen HS, Nardelli B, Murray JH, Zhou JX, Garcia A, Moody G, Zaritskaya LS, Sung C. Pharmacokinetic and pharmacodynamic studies of a human serum albumin-interferon-alpha fusion protein in cynomolgus monkeys. J Pharmacol Exp Ther. 2002;303:540-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 160] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 25. | Feng GS, Gray PW, Shepard HM, Taylor MW. Antiproliferative activity of a hybrid protein between interferon-gamma and tumor necrosis factor-beta. Science. 1988;241:1501-1503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 26. | McCormick AL, Thomas MS, Heath AW. Immunization with an interferon-gamma-gp120 fusion protein induces enhanced immune responses to human immunodeficiency virus gp120. J Infect Dis. 2001;184:1423-1430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 27. | Kreitman RJ. Toxin-labeled monoclonal antibodies. Curr Pharm Biotechnol. 2001;2:313-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 37] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 28. | Slodowski O, Böhm J, Schöne B, Otto B. Carboxy-terminal truncated rhuIFN-gamma with a substitution of Gln133 or Ser132 to leucine leads to higher biological activity than in the wild type. Eur J Biochem. 1991;202:1133-1140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
Science Editor Li WZ Language Editor Elsevier HK