Published online Apr 28, 2005. doi: 10.3748/wjg.v11.i16.2482
Revised: March 14, 2004
Accepted: April 7, 2004
Published online: April 28, 2005
AIM: To investigate the expression of cellular FLICE (Fas associated death domain-like IL-1beta-converting enzyme)-inhibitory protein (c-FLIP) and its association with p53 mutation in colon cancer.
METHODS: Immunohistochemical staining of c-FLIP and mutant p53 by using specific antibodies was performed by the standard streptavidin-peroxidase technique for 45 colon cancer tissue samples with matched normal tissues. Semi-quantitative reverse transcriptional (RT)-PCR was used to measure c-FLIP mRNA levels. t-test statistical method was used in data analyses.
RESULTS: c-FLIP mRNA was expressed in all colon cancer tissues and its level (0.63±0.12) was significantly higher than that in normal tissues (0.38±0.10, P<0.01). Immuno-histochemically, c-FLIP protein was also expressed in all colon cancers (45/45) and 71.1% (32/45) showed an intense immunostaining, in contrast, 93.3% (42/45) of normal colonic mucosa showed positive staining and none of them immunostained intensely. The quantity of c-FLIP protein was significantly higher in cancer tissues than in normal mucosa (7.04±1.20 vs 5.21±0.86, P<0.01). Positive staining of mutant p53 protein was found in 60% (27/45) colon cancers. c-FLIP mRNA level was decreased in p53 positive group compared with p53 negative cancer tissues (0.59±0.13 vs 0.69±0.14, P<0.01), but c-FLIP protein had a significantly higher level in p53 positive cancer tissues than in negative ones (7.57±1.30 vs 6.25±1.27, P<0.01).
CONCLUSION: c-FLIP is specially overexpressed in colon cancers and it might contribute to carcinogenesis of normal colonic mucosa. p53 may exert transcriptional upregulation effects on c-FLIP gene and more potent effects on promoting the degradation of c-FLIP protein.
- Citation: Zhou XD, Yu JP, Chen HX, Yu HG, Luo HS. Expression of cellular FLICE-inhibitory protein and its association with p53 mutation in colon cancer. World J Gastroenterol 2005; 11(16): 2482-2485
- URL: https://www.wjgnet.com/1007-9327/full/v11/i16/2482.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i16.2482
The development and progression of colorectal cancer involves unregulated epithelial cell proliferation associated with a series of accumulated genetic alterations. There is evidence that prolonged survival of such genetically unstable colonic epithelial cells, with their ultimate malignant transformation, is associated with progressive inhibition of apoptosis. Apoptosis is a morphologically distinct form of cell death, which is genetically regulated and, in addition to other roles, provides a vital protective mechanism against the development of neoplasia by removing cells with DNA damage. Inhibition of apoptosis thus confers a survival advantage on cell harboring genetic alterations and may promote acquisition of further mutations to cause neoplastic progression, and also contribute to the development of resistance to chemotherapy.
A new class of virus-encoded apoptosis inhibitory molecules, designated viral FLICE (Fas associated death domain-like IL-1beta-converting enzyme)-inhibitory protein (v-FLIP), was described previously[1-3]. These molecules are composed of two DEDs (death effect domain)[1]. Cellular homologs of v-FLIP have been identified by different groups and have been termed cellular FLICE-inhibitory protein (c-FLIP), CASH, Casper, CLARP, FLAME-1, I-FLICE, MRIT, and Usurpin. At mRNA level, c-FLIP exists as multiple splice variants, but at protein level only two endogenous forms, c-FLIPlong (c-FLIPL) and c-FLIPshort (c-FLIPS) could be detected so far[4-6]. c-FLIPL is a 55 ku protein and structurally similar to procaspase-8; it contains two DEDs and a caspase-like domain. This domain lacks residues that are important for its catalytic activity, most notably a cysteine residue within the active site. The short form of c-FLIP is a 28 ku protein and structurally resembles v-FLIP containing also two DEDs. Both c-FLIP species were found to be recruited to the DISC (death inducing signaling complex)[6]. A number of studies support the notion that both forms of c-FLIP can prevent Fas/CD95 and other death receptors mediated apoptosis by interacting with either FADD (Fas associated death domain) and/or procaspase-8[2,4,7]. Overexpression of c-FLIP gene was found in many malignancies and played a pivotal role in apoptotic resistance of tumor cells and contributed to tumorigenesis eventually[8-10]. However, the exact in vivo expression status of c-FLIP and its association with p53 mutation in colon cancer have not been characterized until now. In the present study, we performed an expression analysis of c-FLIP at mRNA and protein levels to determine its prevalence in colonic carcinomas and its association with p53 mutation.
Forty-five fresh colon cancer tissues with matched normal tissues from patients with disparate pathological stages were collected and fresh-frozen in liquid nitrogen after surgical resections performed at Renmin Hospital, Wuhan University (Wuhan, China) from 2000 to 2002. Of these, 26 were male and 19 were female. The mean age was 57 years (SD 24.1 years, range, 30-81 years). None of the patients had received chemo-, radio- or immuno-therapy before resection. Part of each specimen was routinely processed, fixed in 40 g/L buffered formalin, and embedded in paraffin for histopathological analysis (hematoxylin and eosin stain) and immunohistoc-hemical staining. All tissues were scored by two pathologists blinded to culture results and disease status. Among the 45 cases, 16 were well-differentiated adenocarcinomas, 27 were moderately differentiated carcinomas, and only 2 were poorly differentiated carcinomas.
Immunohistochemistry Immunohistochemistry streptavidin-peroxidase (S-P) technique was used to detect c-FLIP and mutant p53 protein. Affinity purified rabbit polyclonal anti-human c-FLIP specific IgG and mouse monoclonal anti-human p53 specific IgG recognizing mutant p53 were purchased from Santa Cruz Biotechnology and Neomarkers Biotechnical Company, respectively. Immunostaining S-P kit was purchased from Beijing Zhongshan Biotechnical Company. Immunohistochemistry was performed as follows: Formalin-fixed, paraffin-embedded tissue blocks were serially sectioned at 4 μm. Sections were deparaffinized in xylene and rehydrated before analysis. Endogenous peroxidase was quenched with 3.0% hydrogen peroxide in methanol for 10 min; antigen retrieval was performed by microwave for 15 min and tissue sections were then blocked for 20 min with normal rabbit serum. This was followed by incubation overnight at 4 °C with primary antibody at a dilution of 1:120 for c-FLIP or 1:100 for p53. Incubation with PBS instead of the primary antibody served as a negative control. Sections were washed thrice with PBS for 2 min each and incubated with second antibody for 30 min at room temperature. After washing thrice with PBS for 2 min each, sections were stained by a streptavidin-peroxidase detection system. Antibody binding was visualized using diaminobenzidine as chromogen and counterstained with hematoxylin.
The degree of c-FLIP staining was estimated by semi-quantitative evaluation and categorized by the extent and intensity of staining as follows: (1) The extent of positive cells was estimated as 0: positive staining cells <5%, 1: positive staining cells 5-25%, 2: positive staining cells 26-50%, 3: positive staining cells 51-75%, 4: positive staining cells >75%. (2) The intensity of staining was scored as 0: achromatic, 1: light yellow, 2: yellow, 3: brown. The percentage of positive tumor cells and staining intensity were multiplied to produce a weighted score for each case. Cases with weighted scores <1 were defined as negative; otherwise were defined as positive. p53 staining was graded into two grades: positive cells<5% was defined as negative and ≥5% was defined as positive.
Semi-quantitative reverse transcriptional (RT)-PCR After homogenization, total RNAs were extracted from 45 fresh primary colon cancer tissues and paired normal colonic mucosa, using TRIzol reagent (Invitrogen, San Diego, CA). First strand cDNA was synthesized from 1 µg of total RNA using oligo-dT primer and Moloney murine leukemia virus reverse transcriptase (Promega, Southampton, UK) according to the instructions of the manufacturers. For PCR, the primer sequences and expected product sizes were as follows: c-FLIPL/S, (sense) 5’-TGTTGCTATAGATGTGG-3’ and (antisense) 5’- AAGGATCCTTGAGACTCT-3’, 512 bp; and β-actin, (sense) 5’-TGACGGGGTCACCCACACTGTGCC-3’, (antisense) 5’-CTGCATCCTGTCGGCAATGCCAG-3’, 475 bp. The reaction was performed at 95 °C for 2 min, followed by 35 cycles of denaturing at 95 °C for 45 s, annealing at 55 °C for 45 s and extension at 72 °C for 1 min.
The PCR products were analyzed on 2% agarose gels and visualized by ethidium bromide staining. Quantitation of expression levels was achieved after adjustment for the expression levels of the housekeeping gene β-actin by densitometry (Bio-Rad, Hercules, CA, USA). The relative level of expression was then represented as the ratio of c-FLIPL/S/β-actin in normal tissues and carcinomas.
The statistical software package SPSS 10.0 was used and data were presented as mean±SD. The prevalence of c-FLIP gene expression in cancers and normal tissues and its association with p53 mutation were compared by the Student’s t-test. A P value less than 0.05 was considered to indicate statistical significance.
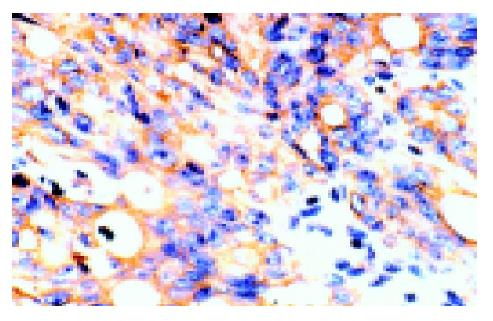
By immunohistochemistry, anti-c-FLIP pAb reacted with colonic mucosal epithelial cells of normal tissues and the majorities were light yellow or yellow micro-granules located in the cytoplasm and/or plasma membranes. In colon cancer, the positive staining showed yellow or brown coloration in the cytoplasm and/or plasma membranes (Figure 1). c-FLIP-positive and -negative areas were found within the same tissue, and the intensity of positive staining was also found to vary within a case tested. After multiplying the weighted c-FLIP scores, the mean expression scores in normal mucosa ranged from 0 to 9 (5.21±0.86); none of them showed a brown immunostaining and 6.7% (3/45) showed negative reactivity. c-FLIP protein was constitutively expressed in all carcinomas (45/45) and the mean scores ranged from 1 to 12 (7.04±1.20); 71.1% (32/45) of them were brown immunostained. Moreover, its level was significantly higher than that of matched normal colonic mucosa (Table 1), (P<0.01).
| Group | Number | c-FLIP protein | P | c-FLIP mRNA | P |
| Normal tissue | 45 | 5.21±0.86 | 0.38±0.10 | ||
| Cancer tissue | 45 | 7.04±1.20 | <0.01 | 0.63±0.12 | <0.01 |
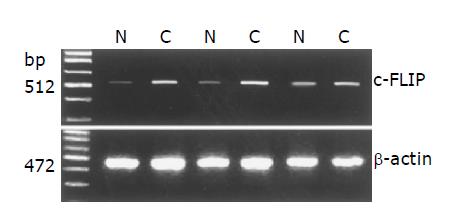
We examined mean expression levels of c-FLIP mRNA in 45 colonic carcinomas and their matched normal tissues, using a semi-quantitative RT-PCR assay. RT-PCR was controlled by equalization of input RNA for each sample and comparable amplification efficiencies were validated by the uniformity of control β-actin RT-PCR product yields. RT-PCR results showed that the adjacent non-tumor specimens and colon cancer specimens constitutively expressed c-FLIP mRNA (Figure 2), but the mean expression levels varied between these tissues. While the mean expression levels of c-FLIP mRNA (c-FLIP/β-actin) in 45 normal colonic tissues ranged from 0.14 to 0.58 (0.38±0.10), the levels in 45 carcinomas ranged from 0.40 to 0.83 (0.63±0.12) (Table 1). Mean c-FLIP mRNA expression in carcinoma was significantly higher than in normal colonic mucosa (P<0.01).
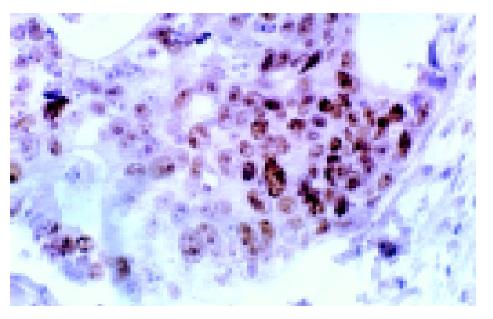
Immunohistochemical results showed that mutant p53 was expressed in 60% (27/45) colon cancer tissues with positive staining in the nuclei of tumor cells (Figure 3). To determine the relationship between p53 protein and c-FLIP level, we compared the expression level of c-FLIP in p53 positive cancer group with that in p53 negative group. As a result (Table 2), c-FLIP mRNA level was decreased in p53 positive cancer group compared with p53 negative cancer tissues (0.59±0.13 vs 0.69±0.14, P<0.01), but c-FLIP protein had a significantly higher level in p53 positive cancer tissues than in negative ones (7.57±1.30 vs 6.25±1.27, P<0.01).
| Group | Number | c-FLIP protein | P | c-FLIP mRNA | P |
| p53 positive | 27 | 7.57±1.30 | 0.59±0.13 | ||
| p53 negative | 18 | 6.25±1.27 | <0.01 | 0.69±0.14 | <0.01 |
In the last decade, basic cancer research has focused on the deregulation of apoptosis as a central event in the process of carcinogenesis and a number of studies showed a central role of apoptosis in tumorigenesis of colon cancer. Varied regulators of apoptosis including p53, survivin, Fas and nuclear factor-kappa B have been implicated in the development of colon cancer[11]. A novel anti-apoptotic gene, designated c-FLIP, has been demonstrated to be overexpressed in several human cancers, suggesting an important role of c-FLIP in cancer development[8-10]. However, the exact expression status of c-FLIP and its possible regulation mechanisms in colon cancer have not been determined.
In this study, we detected the expression level of c-FLIP gene in colon cancer by using immunohistochemistry and RT-PCR. The results showed that the expression of c-FLIP gene was frequently upregulated at both mRNA and protein levels in colonic carcinoma compared with their normal counterparts, suggesting that increased expression of c-FLIP gene was a tumor specific phenomenon and overexpression of c-FLIP might contribute to tumorigenesis of colon cancer by abrogating death signals of tumor cells conducted by death receptors such as Fas, TNF-R, DR3, TRAIL-R, etc. Recently, Ryu et al[12], also reported that c-FLIP was specially overexpressed in colon cancer with the same positive rates of mRNA and protein as detected in our experiments, but the positive rate of c-FLIP protein in normal colonic mucosa was lower than the present research (89.6% vs 93.3%). We deduced that it was at least partially due to the different antibodies used to detect the protein level in immunostaining. The antibody used in the previous study was designed only to recognize the long form of c-FLIP (c-FLIPL); however, the antibody used in our study was expected to recognize both forms of c-FLIP protein (c-FLIPL and c-FLIPS), both of which were demonstrated to act as inhibitors of apoptosis[1,4,7].
Wild type (wt) p53 gene is a tumor suppressor gene, playing an important role in inducing cell cycle arrest and apoptosis. Wt p53 gene mutation is one of the most common genomic alterations in human neoplasia. Because of its short and unstable half-life, it cannot be detected by immunohistochemical examinations; mutant p53 gene product, a more stable protein product accumulated in cells having a loss of its normal function by extending its half-life, may inactivate the wt p53. As we all know, wt p53 is a well-known transcriptional activator of target gene and can induce apoptosis depending on the transcriptional regulation of apoptosis-related genes. Bartke et al, systematically analyzed mRNA level of about 60 apoptosis-related genes by means of RNAse protection assays and found that 15% of these genes were significantly regulated by wt p53. Furthermore, the transcriptional regulation of wt p53 target genes was based on a common principle, in which pro-apoptotic factors were upregulated and anti-apoptotic factors were repressed by p53. However, c-FLIP was found to be a remarkable exception as its mRNA was upregulated by wt p53 in colon cancer cell lines[13]. Still little is known about the relationship between p53 and c-FLIP in vivo. At the beginning of the current study, we supposed that c-FLIP mRNA and protein levels would be downregulated accompanied with p53 mutation. As a result, c-FLIP mRNA level decreased in mutant p53 positive cancer group, whereas c-FLIP protein level was significantly upregulated unexpectedly. It seems this is a contradictory result. We speculated other mechanisms might act on the regulation of c-FLIP protein level besides transcriptional upregulation mechanism. Fukazawa et al[14], have recently shown that wt p53 could enhance the degradation of FLIP via a ubiquitin-proteasome pathway resulting in sensitization of human colon cancer cells to apoptosis. Taken together, we deduced that wt p53 could also enhance the degradation of c-FLIP protein in vivo and this effect might be more potent than the transcriptional upregulation of c-FLIP gene. Thus, c-FLIP protein level elevated as a result of dramatically decreased degradation in spite of relatively moderate downregulation of c-FLIP mRNA when p53 mutated in colon cancer. Further studies are needed to validate it.
In summary, our data show that c-FLIP is overexpressed in colon cancer than in matched normal tissues and may promote the evasion of tumor cells from apoptosis that eventually contribute to carcinogenesis and/or progression of the tumor. p53 may exert transcriptional upregulation effects on c-FLIP gene in vivo and also a more potent effect on promoting the degradation of c-FLIP protein in the mean time.
| 1. | Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer JL, Schröter M. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1029] [Cited by in RCA: 993] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 2. | Hu S, Vincenz C, Ni J, Gentz R, Dixit VM. I-FLICE, a novel inhibitor of tumor necrosis factor receptor-1- and CD-95-induced apoptosis. J Biol Chem. 1997;272:17255-17257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 329] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 3. | Bertin J, Armstrong RC, Ottilie S, Martin DA, Wang Y, Banks S, Wang GH, Senkevich TG, Alnemri ES, Moss B. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:1172-1176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 338] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 4. | Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer JL, Schröter M, Burns K, Mattmann C. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1947] [Cited by in RCA: 1930] [Article Influence: 66.6] [Reference Citation Analysis (7)] |
| 5. | Shu HB, Halpin DR, Goeddel DV. Casper is a FADD- and caspase-related inducer of apoptosis. Immunity. 1997;6:751-763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 316] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 6. | Scaffidi C, Schmitz I, Krammer PH, Peter ME. The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem. 1999;274:1541-1548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 617] [Cited by in RCA: 622] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 7. | Xiao C, Yang BF, Asadi N, Beguinot F, Hao C. Tumor necrosis factor-related apoptosis-inducing ligand-induced death-inducing signaling complex and its modulation by c-FLIP and PED/PEA-15 in glioma cells. J Biol Chem. 2002;277:25020-25025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 146] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 8. | Bullani RR, Huard B, Viard-Leveugle I, Byers HR, Irmler M, Saurat JH, Tschopp J, French LE. Selective expression of FLIP in malignant melanocytic skin lesions. J Invest Dermatol. 2001;117:360-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 81] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Thomas RK, Kallenborn A, Wickenhauser C, Schultze JL, Draube A, Vockerodt M, Re D, Diehl V, Wolf J. Constitutive expression of c-FLIP in Hodgkin and Reed-Sternberg cells. Am J Pathol. 2002;160:1521-1528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 72] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Zhou XD, Yu JP, Liu J, Luo HS, Chen HX, Yu HG. Overexpression of cellular FLICE-inhibitory protein (FLIP) in gastric adenocarcinoma. Clin Sci (Lond). 2004;106:397-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 54] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 11. | Yu LL, Yu JP, Ran ZX, Yu HG. Relationship between nuclear factor-kappa B, apoptosis and proliferation in colorectal neoplasia. Shijie Huaren Xiaohua Zazhi. 2002;10:309-312. |
| 12. | Ryu BK, Lee MG, Chi SG, Kim YW, Park JH. Increased expression of cFLIP(L) in colonic adenocarcinoma. J Pathol. 2001;194:15-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 92] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | Bartke T, Siegmund D, Peters N, Reichwein M, Henkler F, Scheurich P, Wajant H. p53 upregulates cFLIP, inhibits transcription of NF-kappaB-regulated genes and induces caspase-8-independent cell death in DLD-1 cells. Oncogene. 2001;20:571-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 80] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 14. | Fukazawa T, Fujiwara T, Uno F, Teraishi F, Kadowaki Y, Itoshima T, Takata Y, Kagawa S, Roth JA, Tschopp J. Accelerated degradation of cellular FLIP protein through the ubiquitin-proteasome pathway in p53-mediated apoptosis of human cancer cells. Oncogene. 2001;20:5225-5231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 114] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
Co-first-authors: Xiao-Dong Zhou and Hong-Xia Chen
Science Editor Zhu LH Language Editor Elsevier HK