Published online Apr 21, 2005. doi: 10.3748/wjg.v11.i15.2260
Revised: October 16, 2003
Accepted: December 16, 2003
Published online: April 21, 2005
AIM: To construct a recombinant strain which expresses adhesin AlpA of Helicobacter pylori (H pylori ) and to study the immunogenicity of adhesin AlpA.
METHODS: Gene Ab, which was amplified from H pylori chromosomal DNA by PCR technique, was sequenced and the biological information was analyzed, and inserted into the Nco I and Not I restriction fragments of the expression vector pET-22b(+) using T4 DNA ligase. The resulting plasmid pET-AlpA was transformed into competent E.coli BL21(DE3) cells using ampicillin resistance for selection. Recombinant strains were incubated in 5 mL LB with 100 μg/mL ampicillin overnight at 37 °C. Sonication of BL21(DE3)pET-22b(+)/AlpA was analyzed by Western blot to detect AlpA immunogenicity.
RESULTS: The gene encoding AlpA protein was amplified by PCR with chromosomal DNA of H pylori Sydney strain (SS1) as templates. It revealed that AlpA DNA fragment amplified by PCR had approximately 1 500 nucleotides, compatible with the previous reports. The recombinant plasmid pET-22b(+)/AB was successfully constructed. DNA sequencing showed one open reading frame with the length of 588 bp. It encoded seven conservative regions that showed good antigenicity and hydrophobicity by Parker and Welling method. Furthermore, INTERNET EXPASY, NNPREDICT and ISREC predicted that it was a porin-like structure consisting of β-pleated sheets that were embedded in the outer membrane. BLAST analyzed 836 767 protein sequences and found that the similar sequences were all belonging to H pylori OMP sequences. SDS-PAGE and scan analysis showed that the molecular weight of AB was 22.5 ku and recombinant protein amounted to 29% of the total bacterial protein, among which dissolved expression amounted to 21.9% of sonicated supernatant. The rAB purity amounted to 96% through affinity chromatography. Western blot analysis of rAB confirmed that it could be specially recognized by serum form rabbit immunized with AlpA and H pylori infected.
CONCLUSION: Adhesin AlpA recombinant protein may be a potential vaccine for control and treatment of H pylori infection.
-
Citation: Xue J, Bai Y, Chen Y, Wang JD, Zhang ZS, Zhang YL, Zhou DY. Expression of
Helicobacter pylori AlpA protein and its immunogenicity. World J Gastroenterol 2005; 11(15): 2260-2263 - URL: https://www.wjgnet.com/1007-9327/full/v11/i15/2260.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i15.2260
Helicobacter pylori (H pylori) infection is the major cause of chronic active gastritis and peptic ulcer disease[1], and is also closely related with gastric cancers such as adenocarcinomas, mucosa-associated lymphoid tissue lymphoma and primary gastric non-Hodgkin’s lymphoma[2]. This organism has been categorized as a class I carcinogen by the World Health Organization, and direct evidence of its carcinogenicity was recently demonstrated in China. In addition, seroprevalence studies indicate that H pylori infection is also associated with cardiovascular, respiratory, extra-gastroduodenal digestive, autoimmune diseases. Successful eradication of H pylori is thus an important goal. Currently, treatment involves antibiotic therapy, but this has associated problems such as low patient compliance and an increase of resistant strains. An alternative approach is to develop a vaccine, which would not only clear the organism, but also protect against reinfection.
In the development of H pylori vaccine, the candidate vaccine antigens adopted currently such as urease, vacuolating cytotoxin and catalase[3] focused basically on blocking toxicity factors of H pylori, while few candidate vaccine antigens focused on adhesins, which are closely associated with H pylori colonizing in human gastric mucosa by adhering to mucous epithelial cells and the mucus layer lining the gastric epithelium. It is probably a useful attempt to discover protective antigens based on adhesin. Currently, four adhesins have been recognized[4-8], including H pylori adhesin BabA[9,10] whose receptor has been determined till date and other three adhesins of AlpA[11], AlpB[11] and HopZ[12]. No concerned study has been reported about AlpA in China. So in this study, the recombinant plasmid of H pylori AlpA gene was constructed for development of H pylori vaccine.
Bacterial strain BL21(DE3) and plasmid pET-22b(+) were provided by the Institute of Biotechnology, Academy of Military Medical Sciences. H pylori SS1 was preserved in this research institute. Restriction enzymes Not I, Nco I and T4 DNA ligase, Vent DNA polymerase and isopropyl-β-D-thiogalactopyranoside (IPTG) were purchased from New England Biolabs. Goat anti-rabbit and goat anti-human IgG-HRP were purchased from Huamei Bioengineering Company, China, and His-Tag precolumn from Invitrogen. The serum from rabbits immunized with AlpA was donated by Odenbreit et al[11]. Serum samples were obtained from H pylori positive and negative patients who underwent urease test, pathologic examination and germiculture at the Endoscope Center of this institute. Other reagents were analytically pure reagents produced in China.
All restriction enzyme digestions, ligations and other common DNA manipulations, unless otherwise stated, were performed by standard procedures[13]. The genome of H pylori was prepared from cells collected from the colonies on the agar plate. The gene of H pylori AlpA was amplified from the genome of H pylori by PCR (Techne PROGENE) using primers AlpA1 (5’- TGGCCATGGATTGCGCTAGCATAAGTTA -3’) as upstream primer and AlpA2 (5’- AGTGCGGCCGCGAATGAATACCCATAAGA -3’) as downstream primer as described in the literature[11]. AlpA1 and AlpA2 contained Nco I and Not I sites, respectively. PCR was performed with the hot start method. The PCR condition was initial denaturing at 95 °C for 30 s, each cycle of amplification consisted of denaturing at 95 °C for 30 s, annealing at 55 °C for 30 s and polymerization at 72 °C for 50 s and further polymerization for 10 min after 35 PCR cycles. The PCR products were harvested from agarose gel, digested with Nco I and Not I, and inserted into the Nco I and Not I restriction fragments of the expression vector pET-22b(+) using T4 DNA ligase. The resulting plasmid pET-AlpA was transformed into competent E.coli BL21(DE3) cells using ampicillin resistance for selection. The alkaline lysis method was used for large-scale preparations of the recombinant plasmid and the plasmid was identified by restriction enzymes. DNA sequence was performed with the DNA automatic sequencer.
Recombinant strains were incubated in 5 mL LB with 100 μg/mL ampicillin overnight at 37 °C. Fifty microliters LB was inoculated and the cells grew until the optical density at 600 nm reached 0.4-0.6. IPTG was added to the cultures at the final concentrations of 0.1, 0.2, 0.4, 0.6, 0.8 and 1.0 mmol/L, respectively. E.coli cells were harvested after 3 or 5 h by centrifugation at 12000 g for 10 min. The pellet was resuspended in 1 mL 30 mmol/L Tris buffer (pH 8.0) containing 1 mmol/L EDTA (pH 8.0) and 20% sucrose. The suspension was put on ice for 10 min, then centrifuged for 10 min at 12000 g, and the resulting supernatant contained proteins from the periplasm. The resulting pellet was resuspended in 5 mL 50 mmol/L Tris buffer (pH 8.0) containing 2 mmol/L EDTA, 0.1 mg/mL lysozyme and 1% Triton X-100. The suspension was incubated at 30 °C for 20 min and then sonicated on ice until it became clarified. The lysate was centrifuged at 12000 g for 15 min at 4 °C. Whole-cell lysates, sonicated supernatant, inclusion body, osmotic shock liquid of recombinant strains expressing H pylori AlpA were analyzed by electrophoresis in a 10% polyacrylamide gel.
Sonication of BL21(DE3)pET-22b(+)/AlpA was analyzed by Western blot to detect AlpA immunogenicity. Protein was separated on 10% SDS-PAGE gel using a mini-gel apparatus (Bio-Rad) and transferred to nitrocellulose membranes using a semi-dry blot system (Biotech Fischer). The polyclonal rabbit antiserum and sera from patients infected with H pylori were pre-absorbed with BL21(DE3)pET-22b(+) sonication, to remove non-specific antibodies diluted at 1:50. The membrane was incubated for 90 min at 37 °C, washed thrice for 15 min with TBS/Tween-20 (TTBS), and then incubated for 1 h with goat-anti rabbit IgG alkaline phosphates conjugate at room temperature. After washing thrice with TTBS, BCIP/NBT was used to visualize bound antibodies.
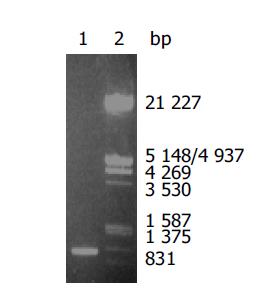
The gene encoding AlpA protein was amplified by PCR with chromosomal DNA of H pylori Sydney strain (SS1) as templates. The cloned products were electrophoresed and visualized on 8 g/L agarose gel (Figure 1). It revealed that AlpA DNA fragment amplified by PCR had approximately 1500 nucleotides, compatible with the previous reports[11].
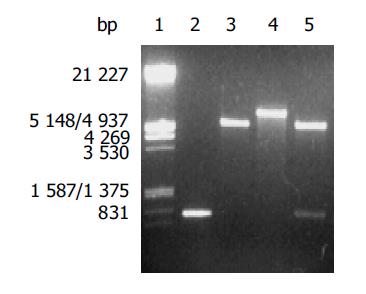
After the PCR products and pET-22b(+) plasmid were cut by Not I and Nco I, directional cloning was performed, resulting in a recombinant plasmid named pET-22b(+)/AlpA. The recombinant plasmids pET-22b(+)/AlpA were all digested by Not I or Nco I, and by Not I and Nco I simultaneously, and then the digestive products were visualized on 8 g/L agarose gel electrophoreses (Figure 2). It demonstrated that recombinant plasmid contained the objective gene.
The nucleotide sequence of cloned genes inserted in pET22b(+) was analyzed by automated sequencing across the cloning junction, using the universal primer T7. The cloned genes contained 1 512 nucleotides coding a putative protein consisted of 504 amino acid residues with a calculated molecular mass of AlpA. The homogenicity was 97.3% between them.
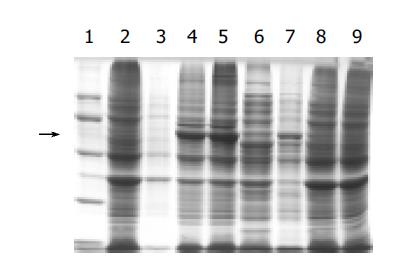
Whole-cell sonication, sonicated supernatant, osmotic shock liquid of recombinant strains expressing H pylori AlpA genes were analyzed by electrophoresis in a 10% polyacrylamide gel for detection of fusion proteins (Figure 3). The result showed that the clearly identifiable band was 56500 ku highly expressed fusion protein, which was similar to that predicted. Gel automatic scan analysis showed that it was 0.6 at D value and the final concentration of IPTG was 0.1 mmol/L after 3-h induction, and the expression of AlpA increased remarkably, which amounted to 31.9% of the total bacterial protein, 23.7% of the sonicated bacterial supernatant and 64.8% of the bacterial inclusion body.
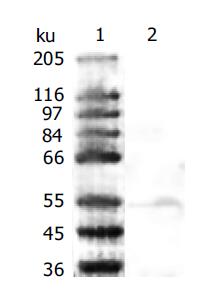
Western blot analysis on the sonicated BL21(DE3)pET-22b(+)/AlpA using polyclonal rabbit antiserum and patients infected with H pylori serum pre-absorbed with sonicated BL21(DE3)pET-22b(+) revealed a protein band, which corresponded to the expected molecular weight (Figure 4). The serum from rabbits without immunization and H pylori negative serum from patients were used as a control. Indeed the control serum did not recognize BL21(DE3)pET-22b(+)/AlpA. It showed that anti-AlpA antibody existed in the serum of patients infected with H pylori and that rAlpA could enable the organism to generate specific humoral immunity.
In 1996, Odenbreit isolated and characterized a chromosomal locus of H pylori previously identified by transposon shuttle mutagenesis as being involved in adhesion of the pathogen to gastric epithelial cells[14]. Afterwards, two close homologous genes were identified, designated as AlpA and AlpB, encoding outer membrane (OM) proteins of 518 amino acids each[11]. They are members of the outer membrane protein supergene family identified in the H pylori 26695 complete genome sequence. Transposon insertion mutagenesis, immunoblotting and primer extension studies indicated that both genes were organized in an operon, but no obvious consensus promoter sequence was found upstream of the transcriptional start site. The C-terminal portion of both proteins was predicted to form a porin-like barrel in the outer membrane, consisting of 14 transmembrane amphipathic strands. Preincubation of H pylori with an antiserum raised against the AlpA fusion protein completely blocked binding of H pylori to the gastric tissue sections, but an antiserum raised against a H pylori urease B fusion protein did not[11]. These data suggested that AlpA might be necessary for specific adhesion of H pylori to human gastric tissues and AlpA must be located on the bacterial surface in order to be bound and functionally be impaired by the specific antiserum. Furthermore, the pattern of AlpA-dependent adherence of H pylori to gastric epithelial surfaces showed a clear difference to the BabA2-mediated adherence to Lewisb, suggesting that a different receptor is involved. Thus, researches suspected that AlpA played an important role in the mechanism of H pylori adherence.
Outer-membrane proteins and porins could form a large family of 32 genes in H pylori. These proteins, which are surface exposed and well conserved, might be excellent vaccine candidates[15,16]. AlpA accords with the criteria mentioned above. Furthermore, AlpA as a candidate component of gene engineering vaccines possess the following virtues. AlpA is a kind of protein so that a gene engineering clone expressing AlpA can be constructed to realize scale-up production and purification. Although AlpA as an adhesion is an important component attending H pylori infection and AlpA itself is lack of toxicity and cross-reactivity with human tissue antigens. AlpA exists in H pylori isolated from multiple geographic locations. These virtues also fit the criteria for inclusion of an antigen in a final vaccine formulation as Kleanthous et al[17], mentioned in 1998. So, to further research, AlpA is helpful in the preparation of H pylori gene engineering vaccines.
AlpA carries a functional lipoprotein signal sequence. When we designed specific primers, we discarded the lipoprotein signal sequence. To facilitate the expression and purification at the next stage, we cloned it into a fusion expression vector with six hercynine tails. The restriction enzyme digestion and sequencing result show one open reading frame with 1512 bp. SDS-PAGE and scanning analysis showed that the molecular weight of AlpA was 56.5 ku and recombinant protein amounted to 31.9% of the total bacterial protein. In this study, the serum from rabbits immunized with purified recombinant protein AlpA could recognize AlpA-specific bands expressed by BL21(DE3)pET-22b(+)/AlpA, but the serum from rabbits in the control group did not. Serum from patients also showed the same results. These results suggest that AlpA of H pylori might be a good candidate as a vaccine component.
| 1. | Wang J, Brooks EG, Bamford KB, Denning TL, Pappo J, Ernst PB. Negative selection of T cells by Helicobacter pylori as a model for bacterial strain selection by immune evasion. J Immunol. 2001;167:926-934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 81] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 2. | Wang J, Fan X, Lindholm C, Bennett M, O'Connoll J, Shanahan F, Brooks EG, Reyes VE, Ernst PB. Helicobacter pylori modulates lymphoepithelial cell interactions leading to epithelial cell damage through Fas/Fas ligand interactions. Infect Immun. 2000;68:4303-4311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 69] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 3. | Bai Y, Zhang YL, Wang JD, Zhang ZS, Zhou DY. Construction of the non-resistant attenuated Salmonella typhimurium strain expressing Helicobacter pylori catalase. Diyi Junyi Daxue Xuebao. 2003;23:101-105. [PubMed] |
| 4. | Bai Y, Zhang YL, Wang JD, Lin HJ, Zhang ZS, Zhou DY. Conservative region of the genes encoding four adhesins of Helicobacter pylori: cloning, sequence analysis and biological information analysis. Diyi Junyi Daxue Xuebao. 2002;22:869-871. [PubMed] |
| 5. | Bai Y, Dan HL, Wang JD, Zhang ZS, Odenbreit S, Zhou DY, Zhang YL. Cloning, expression, purification and identification of conservative region of four Helicobacter pylori adhesin genes in AlpA gene. Prog Biochen Biophys. 2002;29:922-926. |
| 6. | Bai Y, Zhany YL, Chen Y, Wang JD, Zhou DY. Study of Immunogenicity and Safety and Adherence of Conservative Region of Four Helicobacter pylori Adhesin in vitro. Prog Biochen Biophys. 2003;30:422-426. |
| 7. | Bai Y, Zhang YL, Wang JD, Zhang ZS, Zhou DY. Cloning and immunogenicity of conservative region of adhesin gene of Helicobacter pylori. Zhonghua YiXue ZaZhi. 2003;83:736-739. [PubMed] |
| 8. | Bai Y, Wang JD, Zhang ZS, Zhang YL. Construction of the Attenuated Salmonella typhimurium Strain Expressing Helicobacter pylori Conservative Region of Adhesin Antigen. Chin J Biotech. 2003;19:77-82. |
| 9. | Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Borén T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 872] [Cited by in RCA: 850] [Article Influence: 30.4] [Reference Citation Analysis (1)] |
| 10. | Bai Y, Chang SH, Wang JD, Chen Y, Zhang ZS, Zhang YL. Construction of the E.coli clone expressing adhesin BabA of Helicobacter pylori and evaluation of the adherence activity of BabA. Diyi Junyi Daxue Xuebao. 2003;23:293-295, 309. [PubMed] |
| 11. | Odenbreit S, Till M, Hofreuter D, Faller G, Haas R. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol Microbiol. 1999;31:1537-1548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 185] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 12. | Peck B, Ortkamp M, Diehl KD, Hundt E, Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999;27:3325-3333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 133] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 13. | Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual.2nd ed. New York: Cold Spring Harbor Laboratory Press 1989; 35-400. |
| 14. | Odenbreit S, Till M, Haas R. Optimized BlaM-transposon shuttle mutagenesis of Helicobacter pylori allows the identification of novel genetic loci involved in bacterial virulence. Mol Microbiol. 1996;20:361-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 57] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 15. | Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2635] [Cited by in RCA: 2603] [Article Influence: 89.8] [Reference Citation Analysis (0)] |
| 16. | Doig P, Trust TJ. Identification of surface-exposed outer membrane antigens of Helicobacter pylori. Infect Immun. 1994;62:4526-4533. [PubMed] |
| 17. | Kleanthous H, Lee CK, Monath TP. Vaccine development against infection with Helicobacter pylori. Br Med Bull. 1998;54:229-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |