Published online Apr 21, 2005. doi: 10.3748/wjg.v11.i15.2230
Revised: March 27, 2004
Accepted: June 7, 2004
Published online: April 21, 2005
AIM: To investigate the inhibitory effect of specialized human telomerase antisense oligodeoxyribonucleotides on the growth of well (MKN-28), moderately (SGC-7901) and poorly (MKN-45) differentiated gastric cancer cell lines under specific conditions and its inhibition mechanism, and to observe the correlation between the growth inhibition ratio and the tumor pathologic subtype of gastric cancer cells.
METHODS: Telomerase activity in three gastric cancer cell lines of variant tumor pathologic subtype was determined by modified TRAP assay before and after the specialized human telomerase antisense oligodeoxyribonucleotides were dealt with under specific conditions. Effect of antisense oligomer under specific conditions of the growth and viability of gastric cancer cell lines was explored by using trypan blue dye exclusion assay, and cell apoptosis was detected by cell morphology observation, flow cytometry and TUNEL assay.
RESULTS: Telomerase activity was detected in well, moderately and poorly differentiated gastric cancer cell lines (the quantification expression of telomerase activity was 43.7TPG, 56.5TPG, 76.7TPG, respectively).Telomerase activity was controlled to 30.2TPG, 36.3TPG and 35.2TPG for MKN-28, SGC-7901 and MKN-45 cell lines respectively after treatment with human telomerase antisense oligomers at the concentration of 5 μmol/L, and was entirely inhibited at 10 μmol/L, against the template region of telomerase RNA component, whereas no inhibition effect was detected in missense oligomers (P<0.05). After treatment with antisense oligomers at different concentrations under specific conditions for 96 h, significant growth inhibition effects were found in MKN-45 and SGC-7901 gastric cancer cell lines (the inhibition ratio was 40.89% and 71.28%), but not in MKN-28 cell lines (15.86%). The ratio of inactive SGC-7901 cells increased according to the prolongation of treatment from 48 to 96 h. Missense oligomers could not lead to the same effect (P<0.05). Apoptosis of SGC-7901 and MKN-45 cells was detected not only by morphology and TUNEL assay but also by flow cytometry. The apoptotic rate reached 33.56% for SGC-7901 cells and 44.75% for MKN-45 cells.
CONCLUSION: The viability and proliferation of gastric cancer cells can be inhibited by antisense telomerase oligomers. The growth inhibition of gastric cancer cells is correlated with concentration, time and sequence specialty of antisense oligomers. The inhibition mechanism of antisense human telomerase oligomers depends not only on the sequence specialty but also on the biological characteristics of gastric cancer cell lines.
- Citation: Ye J, Wu YL, Zhang S, Chen Z, Guo LX, Zhou RY, Xie H. Inhibitory effect of human telomerase antisense oligodeoxyribonucleotides on the growth of gastric cancer cell lines in variant tumor pathological subtype. World J Gastroenterol 2005; 11(15): 2230-2237
- URL: https://www.wjgnet.com/1007-9327/full/v11/i15/2230.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i15.2230
Telomere, a complex structure present at the ends of eukaryotic chromosomes, is regarded as a mitotic clock, eventually signaling the senescence of programed cell death in aged cells, and thus preventing further accumulated genetic changes and chromosome instability that can lead to pathologic lesions such as cancer. So maintenance of telomeres is essential to the proliferation of tumor cells[1]. Telomerase, a ribonucleoprotein, is actually a reverse transcriptase consisting of RNA and protein, the function of which is to add hexameric repeats of 5’-TTAGGG-3’ to the end of telomeres to compensate for the progressive loss of telomeres[2]. It has been approved that telomerase is an enzyme associated with cellular immortality and tumorigenesis, and strangely repressed in most normal somatic cells, but is reactivated in malignant tumor cells or immortal cells. Without the activation of telomerase, oncogene alone is not sufficient to make human cells become malignant phenotypes[3]. Studies have confirmed that telomerase activity is generally undetectable in normal gastric tissue, but is expressed in approximately 85-100% of human gastric cancers[4]. These facts strongly support the idea that telomerase has a promising potential, not only as a useful predictive diagnostic marker for gastric cancer, but also as a novel targeting approach of gastric cancer therapy.
Gastric cancer is one of the most common malignancies and the major cause of cancer-related death in many countries, including China, where it accounts for 15.4 out of 100000 persons[5]. Much has been done to improve the earlier diagnosis and survival rate of gastric cancer through telomerase activity detection and telomerase activity inhibition by anticancer chemotherapy, but according to MEDLINE searches up to the year of 2004, little is known about the effect of antisense human telomerase oligodeoxyribonucleotides on the growth inhibition of gastric cancer cell lines of variant pathologic differentiations and their inhibition mechanism, and the relationship between the inhibition effect of telomerase antisense oligomers and tumor pathologic subtype. The strategy we performed here was to investigate the inhibitory effect of specialized human telomerase antisense oligodeoxyribonucleotides on the growth of well-, moderately- and poorly-differentiated gastric cancer cell lines under specific conditions and the possible mechanism of antisense oligodeoxyribonucleotides, to observe the correlation between the inhibition ratio and tumor pathologic differentiation of tumor cells, or other factors that might affect the inhibition effect of antisense oligodeoxyribonucleotides.
Human gastric cancer tissue and normal gastric tissue were obtained from Shanghai Ruijin Hospital. MKN-28, the well-differentiated human gastric cancer cells and MKN-45, the poorly-differentiated human gastric cancer cells were kindly provided by Japanese Foundation of Cancer Research, Tokyo, Japan. SGC-7901, the moderately-differentiated human gastric cancer cells were present as a gift from No.6 People’s Hospital, Shanghai, China. All tumor cells were cultured in RPMI-1640 median (Gibco) supplemented with 10% heat-inactivated fetal calf serum at 37 °C in a modified CO2 incubator containing 50 mL/L CO2 and 95% air.
Fifteen-mer specific human telomerase antisense oligodeoxyribonucleotides (AS-ODN, 5’-GTTAGGGTTAGACAA-3’) and missense oligodeoxyribonucleotides (N-ODN, 5’-ACGTCGATCTGAGCA-3’) were synthesized by β-cyanoethyl-phospharamidite chemistry using a model 381, an automated DNA synthesizer (Applied Biosystems). The assay procedure and precautions were in accordance with the instructions. Briefly, each basic group was synthesized including deblocking DMT at a controlled pore glass by trichloroacetic acids, then activated, couple purified, condensed, oxidized and aminolyzed according to the protocol of the user’s manual (Applied Biosystems). Electrophoresis in a polyacrylamide gel under denaturing conditions was used for the purification of oligomers. The oligomers were added to cell culture medium containing stable modifications with phosphorothiodate residues.
A highly sensitive method was developed with some modifications for quantification analysis of telomerase activity according to Zhang et al[6,7] in our research center.
CHAPS lysis buffer: 10 mmol/L Tris–HCl (pH 7.5), 1 mmol/L MgCl2, 1 mmol/L EGTA, 0.1 mmol/L PMSF, 5 mmol/L β-mercaptoethanol, 0.5% CHAPS (Sigma), 10% glycerol. TRAP reaction buffer: 20 mmol/L Tris-HCl (pH 8.3), 1.5 mmol/L MgCl2, 63 mmol/L KCl, 0.005% Tween-20, 1 mmol/L EGTA, 50 μmol/L dNTPs (Promega), 0.1 μg TS primer 5’-AATCCGTCGAGCAGAGTT-3’ (All the primers used were synthesized by Shanghai Department of Sangon, Canada). ACX, the return primer: 5’-GCGCGG[CTTACC]3CTAACC-3’. NT, internal control primer: 5’-ATCGCTTCTCGGCCTTTT-3’. TSNT, internal control template: 5’-AATCCGTCGAGCAGAGTTAAAAGGCCGAGAAGCGAT-3’. R8, the quantification standard of oligodeoxyribonucleotide: 5’-AATCCGTCGAGCAGAGTTAG[GGTTAG]7-3’. Taq DNA polymerase (Promega). SYBR Green I (FCM Bioproducts).
All cells and tissues were washed twice with phosphate-buffered saline (0.05 mol/L, pH 7.0), centrifuged (1000 r/min, 5 min) suspended in ice-cold CHAPS-lysis buffer (0 °C, 30 min), centrifuged once again (BECKMAN, 4 °C, 12000 g, 30 min). The supernatant was rapidly frozen and stored at -70 °C.
The concentration of proteins was measured by using the Bradford protein assay, an aliquot of extract containing 1 μg of proteins was used for each telomerase assay.
One microgram protein of telomerase extracts was added into 50 μL TRAP reaction, after 30 min of incubation at 30 °C, 0.1 μg ACX, 0.1 μg NT 0.003 amol TSNT and 2U Taq DNA polymerase were added. The reaction mixture was then subjected to polymerase chain reaction (PCR) amplification in a thermal cycler (Perkin-Elmer 2400) with 30 cycles at 94 °C for 30 s, at 60 °C for 30 s. The PCR products were resolved by electrophoresis in a 12% polyacrylamide gel under nondenaturing conditions. The gel was then stained with SYBR Green I (FCM bioproducts) for 15 min, visualized by the UVP system and analyzed by Gelworks 1D advanced software. In each experiment, negative control (1 μL CHAPS lysis buffer or 1 μg extracts incubated with 5 g/L RNase for 20 min at 37 °C) and positive control (0.1 amol of R8) were included. Telomerase quantification was calculated with the formulation: TPG={[(TP-B/TI)/[R8-B/RI]]×100}. TP, B, R8, TI and RI were telomerase products in fluorescent counts from the test extract, blank lysis buffer only, R8 quantification standard, and internal controls of the test extract and that of the R8 quantification standard, respectively.
The cytotoxic effect of antisense oligomers on tumor cells was determined using the trypan blue dye exclusion assay. Tumor cells in logarithmic phase were seeded at 104 cells/well (100 μL) in 96-well plates and incubated overnight at 37 °C, then the oligomers were added to the appropriate wells (the final concentrations of 5, 10, 15, 20 and 25 μmol/L were revised for antisense and 10 μmol/L for missense). Cells were harvested after 96 h after administration of oligomers, and cell suspension was diluted to 106/L and cell viability was determined by trypan blue dye.
Then SGC-7901 gastric cancer cell line, the most sensitive to the inhibition effect of antisense oligomers of the three cell lines, was chosen for the next assay to observe whether the cytotoxic effect of antisense oligomers at 48, 72, 96 h was time dependent.
Three gastric cancer cell lines were seeded at 106 cells/L in a six-well microplate. Following a 12-h incubation at 37 °C, 50 mL/L CO2, antisense and missense oligomers were added at a final concentration of 10 μmol/L. Additional controls contained 10 μmol/L missense oligomers and culture medium alone respectively. Cells were harvested 96 h later and resuspended in the solution containing sodium citrate 40 mmol/L, sucrose 250 mmol/L and 5% dimethyl sulfoxide. The suspension was stored at -20 °C for 20 min, then thawed rapidly at room temperature and centrifuged to collect the cells. The cells were then resuspended in a solution containing RNase A (5×104 unit/g, 50 mg/L) and propidium iodide (PI, 20 mg/L). PI-stained cells were analyzed by flow cytometry (Becton-Dickson).
Electron microscope observation was used to determine whether tumor cells displayed an apoptotic morphology. Gastric cancer cells were fixed with 2.5% glutaraldehyde in advance, after being fixed with 2% osmic acid, washed with phosphate-buffered saline, dehydrated in alcohol of different grades, and embedded with Epon 812 to form ultrathin sections. After having dyed in both sodium acetate and lead citrate, the ultrathin sections were observed under an electron microscope.
The increased apoptosis of gastric cancer cells was also evaluated using the TUNEL (TDT-mediated dUTP nick end labeling) assay. DNA fragments were detected with FITC-labeled dUTP and terminal deoxynucleotidyl transferase as described by the supplier of reagents.
Cells (1×105/well) were seeded on four-well plates and treated with oligomers in the same manner as described above at the final concentration of 10 μmol/L for 96 h. Cell samples were air dried. A fresh paraformaldehyde solution (4% in PBS, pH 7.4) was prepared for 1 h at room temperature, slides were rinsed with PBS and incubated in a permeability solution (0.1% Triton X-100, 0.1% sodium citrate) for 2 min on ice, then labeled and analyzed by rinsing slides twice with PBS and adding 50 μL TUNEL reaction mixture to sample. Each experiment should include negative controls containing terminal transferase instead of TUNEL reaction and positive controls containing fixed and permeabilized cells incubated with micrococcal nuclease or DNase I (1 μg/mL in 50 mmol/L Tris-HCl, pH 7.5, 1 mmol/L MgCl2, 1 mg/mL BSA, for 10 min at room temperature) to induce DNA strand breaks. The results were observed and photographed via fluorescence microscopy.
Data were expressed as mean±SD. Comparison between means was tested by Student’s t test, Friedman’s ANOVA and Duncan test in SAS. P<0.05 was considered statistically significant.
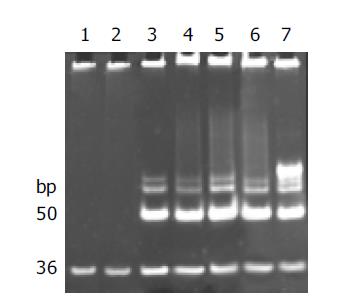
Telomerase activity was detected with the modified TRAP assay. As shown in Figure 1, internal standard bands were detected in all samples starting from 36 bp, by using lysis buffer and normal gastric mucosa as negative controls, human gastric cancer tissue and 0.1 amol R8 as positive controls, which excluded the possibility of false-negative results due to Taq polymerase inhibitor.
Telomerase activity, starting at 50 bp and producing a characteristic 6-bp ladder periodically, was clearly detected in poorly, moderately and well-differentiated gastric cancer cell lines and human gastric cancer tissue, but not detected in normal gastric mucosae. The quantification expression of telomerase activity in poorly-, moderately- and well-differentiated gastric cancer cell lines was MKN-45 76.7TPG, SGC-7901 56.5TPG, MKN-28 43.7TPG. According to the statistical analysis, though the tumor pathologic differentiation and malignancy of these gastric cancer cell lines varied, no significant difference in telomerase activity was observed, which confirmed that the expression of telomerase activity was not correlated with the pathologic differentiation of tumor cells (P>0.05). All of the experiments were repeated for not less than five times.
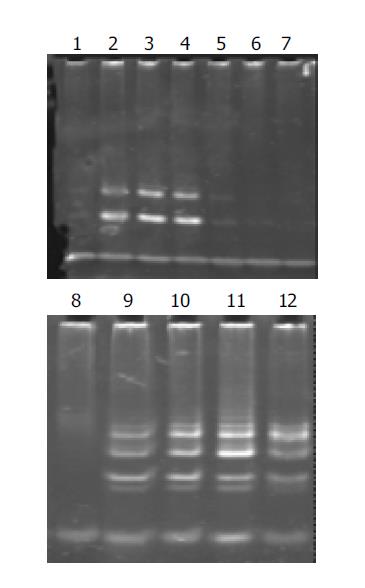
Telomerase activity in well-, moderately-, and poorly-differentiated gastric cancer cell lines was significantly inhibited after having been cultured with antisense human telomerase oligonucleotides for 96 h at the concentrations of 5 and 10 μmol/L, compared to the missense oligomers as contrasts. As shown in Figure 2, telomerase activity was controlled to 30.2TPG, 36.3TPG and 35.2TPG for MKN-28, SGC-7901 and MKN-45 cell lines respectively at the concentration of 5 μmol/L, and was entirely inhibited at 10 μmol/L, while 10 μmol/L missense oligomers could not inhibit telomerase activity.
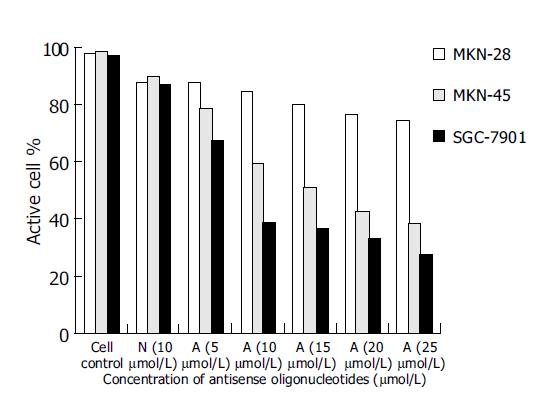
As controls of blank group and missense oligomer group, the antitumor effect of antisense oligomers was remarkable in MKN-45, the poorly-differentiated cells and SGC-7901, the moderately-differentiated cells (P<0.05) at the specified conditions. In both kinds of cell lines, the addition of anticancer agents was associated with concentration-dependent reductions in cell growth inhibition from 5, 10, 15, 20-25 μmol/L. As shown in Figure 3, the slope of curve was a severe slant at the concentration of 10 μmol/L, suggesting that the antitumor effect was even more critical at this point of concentration. Thus, it was a prompt for us to detect the antisense effect on cell apoptosis at this circumstance (10 μmol/L, 96 h). Substantial decrease in cell growth was not seen in MKN-28 groups when analyzed at 96 h (P>0.05).
However, the missense control group did show a decrease in viability (10%), which was not statistically different from the antisense group, and this might represent a nonspecific antisense effect.
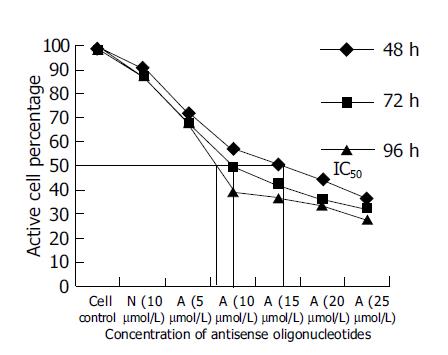
Since SGC-7901, the moderately-differentiated cells, was more sensitive to antitumor effect of antisense oligomers than any other two cell lines, we then chose it to investigate cell viability after incubation with oligomers at 48, 72, 96 h at different concentrations. As shown in Figure 4, when the reaction time was extended from 48 to 96 h, the antisense oligomers displayed a more effective way to inhibit the viability of tumor cells, compared to the missense oligomers as controls (P<0.05). The IC50 of antisense oligomers on growth inhibition of SGC-7901 cancer cells at 48, 72 and 96 h was 15, 10 and 8 μmol/L or so. In this study, the effect of antisense oligomers on gastric cancer cells was not only sequence-specific but also concentration-dependent and time-dependent in cell growth inhibition.
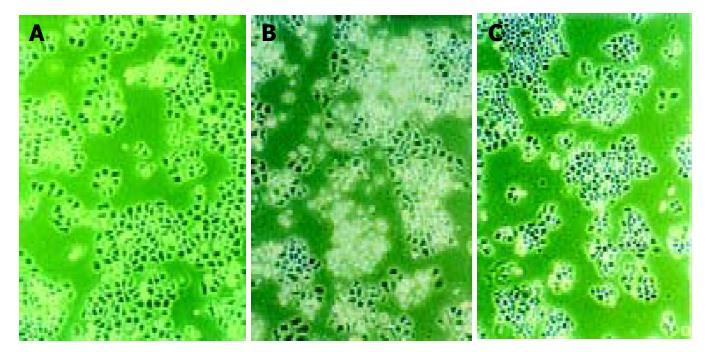
Morphologic observation of cell apoptosis under light microscopy After treatment with 10 μmol/L of antisense oligomers for 96 h, the growth rate of SGC-7901 and MKN-45 declined. When cells stopped proliferating, they showed the morphologic characteristics associated with crisis, such as larger cells with a flattened appearance, membrane blebbing, contact inhibition and adhesion decline (Figure 5), but these changes did not occur to MKN-28, the well-differentiated cells, which had almost the same effect as missense group. Three cell lines in blank group grew exuberantly with bright cell membranes and perfect clear culture solution. All these cells in missense group also grew actively, but not as well as the blank group.
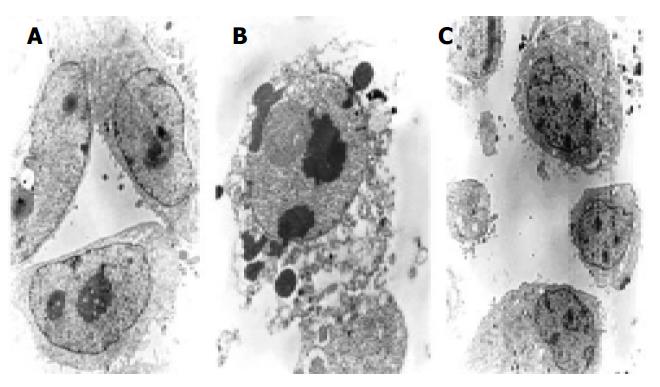
Morphologic observation of cell apoptosis under electron microscopy We investigated whether these cells had the phenotypic hallmarks of apoptosis under electron microscope. Morphologic changes under electron microscope during apoptosis after treatment with 10 μmol/L of antisense oligomers for 96 h were characterized as membrane blebbing instead of microvillus growth, chromatin condensation and margination to the nuclear membrane, nuclear fragmentation and disintegration to form apoptosis corpus (Figure 6). But the MKN-28, the well-differentiated cells, did not have the effect after the same treatment.
Investigating apoptotic rate by flow cytometry To determine whether the cells were undergoing apoptosis, and to examine the apoptotic ratio after treatment of oligomers at 10 μmol/L for 96 h, we performed analysis of cells labeled with PI. Flow cytometric analysis of DNA content demonstrated the accumulation of a sub-G1 peak with the apoptotic rate reaching 33.56% for SGC-7901 cells and 44.75% for MKN-45 cells in antisense group, which was significantly higher than those in missense group (Table 1).
| Group | % Apoptosis | Pa | |
| MKN-28 | Control | 2.68±1.05 | |
| AS-ODN | 17.56±2.57 | P>0.05 | |
| N-ODN | 12.37±4.79 | ||
| SGC-7901 | Control | 2.59±0.94 | |
| AS-ODN | 33.56±3.56 | P<0.05 | |
| N-ODN | 8.46±0.76 | ||
| MKN-45 | Control | 6.77±2.16 | |
| AS-ODN | 44.75±9.93 | P<0.05 | |
| N-ODN | 15.97±4.01 |
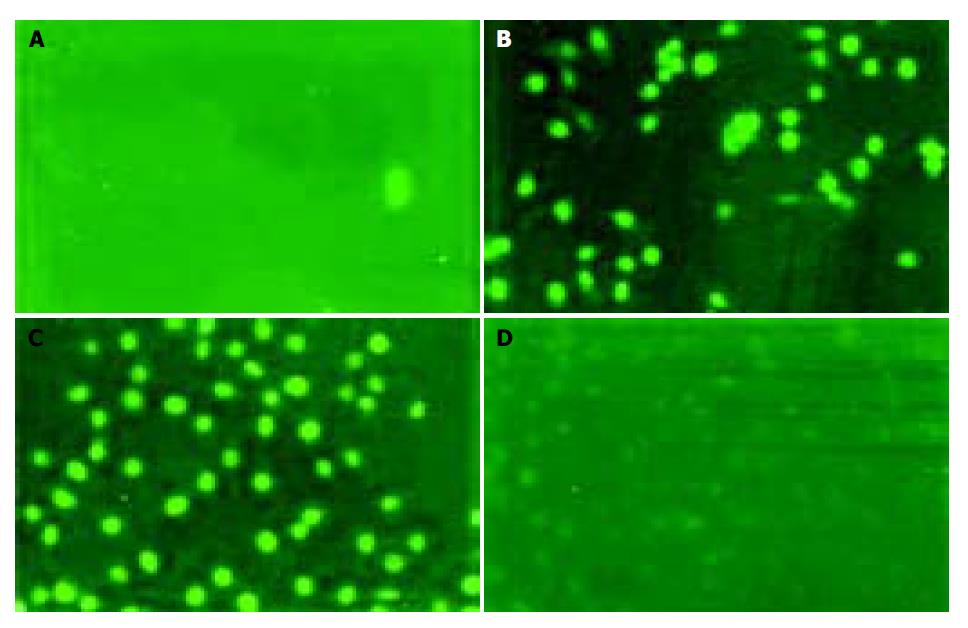
Examining apoptosis by TUNEL assay To further confirm whether gastric cancer cells underwent apoptosis, we performed TUNEL assay. Similar results were obtained by TUNEL assay conforming DNA fragmentation under fluorescence microscopy. Cells in logarithmic phase infiltrating with DNase I (50 μg/mL) were used as a positive contrast and cells in logarithmic phase without any treatment and reagent as a negative contrast, the results showed SGC-7901 and MKN-45 cells of antisense group were fluoresced and turned to green, rounder, larger, and even polykaryon, which was a characteristic sign of apoptosis. Cells in missense group were not found to have changes under fluorescence microscopy (Figure 7).
The antisense human telomerase oligomers used were sequence specific and complementary to telomerase RNA, which is absolutely required for telomerase reverse transcription and therefore a natural target for anti-telomerase agents. Another strongpoint that we designed for the antisense oligmers was modification of DNA oligomers with phosphorothioate (PS) linkages. The modifications of sugar phosphodiester backbone in these molecules were intended to confer certain desirable characteristics of properties, such as superior binding affinity and therefore specificity to the hTR RNA template, intracellular penetration and importantly, to enable the intact delivery to their targets[8]. Compared to other modifications, such as ribozyme, 2’,5’-oligoadenylate[9], peptide nucleotide acid[10], and RNA interference[11], phosphorothioate oligonucleotides are the current gold standard for antisense therapy and have a bright future in clinical study, because they have acceptable physical and chemical properties and show reasonable resistance to nucleases[12].
Our research confirmed that telomerase activity of well-, moderately-, and poorly-differentiated gastric cancer cells was significantly inhibited by antisense oligomers of sequence specificity at 10 μmol/L for 96 h, and the degree of inactivity had no correlation with tumor pathologic differentiation. They had almost consistent expression of telomerase activity before and after the suppression of antisense oligomers, indicating that the suppression mechanism of antisense oligomers is related to the telomerase activity, and antisense oligomers inhibit telomerase and cell growth via sequence specificity against human telomerase RNA components.
In our research, antisense oligomers of sequence specificity inhibited cell proliferation, especially moderately-differentiated gastric cancer cells (SGC-7901), in a concentration- and time-dependent manner. But our findings also had no significant correlation between the reduction in telomerase activity and cell counts or viability. After having been treated with antisense oligomers at the same condition (96 h, 10 μmol/L), telomerase was completely inactivated, but tumor cells were still partially viable, indicating that antisense human telomerase oligomers inhibit telomerase activity earlier and more effective than cell growth, and antisense oligomers inhibit cell proliferation via inactivation of telomerase. Agents that target telomerase, unlike most conventional chemotherapeutic agents, might not induce cytotoxicity immediately after administration[13,14]. Although many tumors maintained short telomeres, complete inhibition of tumor cell proliferation required a continuous cell division until their telomeres reached a critically short length[15-17].
Light and electron microscopy showed that gastric cancer cells appeared to be the characteristic signs of apoptosis and cell growth inhibition after treatment with antisense oligomers. Flow cytometry also confirmed the same outcome as antisense oligomers that induced cell apoptosis by arresting cells at G0/G1, decreasing the percentage of cells at S and G2/M phases, and reducing the cell growth-rate, but the cells in missense group did not have this effect. These data suggest that inhibition of telomerase could enhance cell inviability and apoptosis in some subpopulations without telomerase activity.
However, antisense oligomers could lead to apoptosis of gastric cancer cells (P<0.05, compared to missense group), but the apoptosis ratio was no more than 30-50%, suggesting that other subpopulations during the treatment with antisense oligomers continued to grow and escaped from apoptosis. In agreement with our results, Kondo et al[18] has furthermore attested in their therapeutic experiment of neuroglioma that treatment with antisense human telomerase oligomers could not induce apoptosis of all neuroglioma cells, and surviving cells demonstrated an increased expression of more differentiated states, such as decreased motility, invasive ability and tumorigenicity. So they concerned that inhibition of telomerase in tumor cells could trigger two distinct pathways, apoptosis and differentiation. We also found that antisense human telomerase oligomers could significantly inhibit the growth and viability of gastric cancer cells. The main mechanism of which is the inhibition of telomerase activity and the induction of cell apoptosis. Further study is needed to approve the possible mechanism of cell differentiation.
In this study, we found that antisense oligomers of sequence specificity inhibited efficiently the proliferation of poorly-differentiated gastric cancer cells (MKN-45) and moderately-differentiated gastric cancer cells (SGC-7901) at specific conditions, but the inhibitory action on MKN-28 was restricted and not reasonable. Furthermore, antisense oligmers could control the growth rate of moderately-differentiated gastric cancer cells (SGC-7901) more effective than that of poorly-differentiated gastric cancer cells (MKN-45), indicating that antisense oligomers have the capability of inhibiting cell growth, but they do not enhance or weaken their inhibiting efficiency according to the malignant grades of gastric cancer. These results suggest that the mechanism of the growth inhibition by antisense oligomers has little correlation with malignancy or its pathologic differentiation of gastric cancer.
These data also suggest that the inhibition effect of antisense oligomers on the viability of gastric cancer cell lines is not only correlated well with the inhibition effect of telomerase activity by specifically combining the human telomerase RNA, but also determined by the different biologic traits of gastric cancer cell lines. Since a variety of stimuli are known to induce or suppress apoptosis, we believe that after telomerase activity is inhibited by antisense oligomers of sequence specificity, the unique signal transmission channel is activated in different gastric cancer cell lines, thus starting up a new pass of controlling tumor growth, resulting in some cell types undergoing apoptosis, some types undergoing differentiation and others continuing to grow. It has been demonstrated that tumors undergoing apoptosis via p53-dependent pathways after treatment with antisense telomerase are easily inhibited compared to those via p53-independent pathways after treatment with antitelomerase. MKN-28 cells, unlike most gastric cancer cells (including MKN-45, and SGC-7901), instead of containing wild-type p53 gene, they harbor a missense mutation at codon 257 binding domain, inducing cell apoptosis in a unique way[19]. These studies give some assumption why antisense human telomerase oligomers in our study significantly inhibited cell growth and induced apoptosis in MKN-45 and SGC-7901 gastric cancer cells, but not in MKN-28 cells. So the inhibiting mechanism of antisense telomerase oligomer is not only determined by the sequence specialty, but also correlated with the biologic traits due to the cell lines themselves. Since different cell lines from the same tumor origin may have variant biologic mechanisms in tumorigenesis, they maintain their immortal proliferation in a particular way after overexpression of telomerase activity. Therefore once telomerase activity is inhibited, the response is different to cell apoptosis and growth inhibition.
In agreement with our results, Kondo et al[18]and Taggart et al[20] have confirmed in their studies that the effect of telomerase inhibitors appears in their own way, and is decided by tumor related genes and different cancer cell subtypes. Though telomerase inactivity may lead to cell apoptosis via telomere shortening, it has been proved that a small proportion of tumor cells could synthesize repetitive sequences of telomere to maintain an effective length via selectively stimulating some unknown bypasses[21,22]. It has been found that antisense oligomers could induce apoptosis via upregulating p53, Rb, or induce differentiation via activation of p21 and p27. It has also been approved that treatment with surviving nonapoptotic cells with antisense oligonucleotides against p27 could induce cell death[18]. To confirm our hypothesis, further study is needed to investigate the real mechanism after telomerase inhibition according to cell lines and cell biology.
In conclusion, antisense oligomers targeting on telomerase RNA can inhibit cell immortalization and proliferation of well-, moderately- and poorly-differentiated gastric cell lines. The main mechanism of which is the specific inactivity of telomerase, and induction of cell apoptosis.
| 1. | Greenwood MJ, Lansdorp PM. Telomeres, telomerase, and hematopoietic stem cell biology. Arch Med Res. 2003;34:489-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 2. | Karayan-Tapon L, Menet E, Guilhot J, Levillain P, Larsen CJ, Kraimps JL. Topoisomerase II alpha and telomerase expression in papillary thyroid carcinomas. Eur J Surg Oncol. 2004;30:73-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 3. | Shen ZY, Xu LY, Li EM, Cai WJ, Chen MH, Shen J, Zeng Y. Telomere and telomerase in the initial stage of immortalization of esophageal epithelial cell. World J Gastroenterol. 2002;8:357-362. [PubMed] |
| 4. | Usselmann B, Newbold M, Morris AG, Nwokolo CU. Telomerase activity and patient survival after surgery for gastric and oesophageal cancer. Eur J Gastroenterol Hepatol. 2001;13:903-908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Tang DQ. Oncology of the Time. 2nd edition. Shanghai: Shanghai Medical University 2000; 695. |
| 6. | Zhang RG, Wang XW, Yuan JH, Guo LX, Xie H. Using a non-radioisotopic, quantitative TRAP-based method detecting telomerase activities in human hepatoma cells. Cell Res. 2000;10:71-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 7. | Zhang RG, Wang XW, Yuan JH, Xie H. Human hepatoma cell telomerase activity inhibition and cell cycle modulation by its RNA component antisense oligodeoxyribonucleotides. Acta Pharmacol Sin. 2000;21:742-746. [PubMed] |
| 8. | Wang ES, Wu K, Chin AC, Chen-Kiang S, Pongracz K, Gryaznov S, Moore MA. Telomerase inhibition with an oligonucleotide telomerase template antagonist: in vitro and in vivo studies in multiple myeloma and lymphoma. Blood. 2004;103:258-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 56] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 9. | Yatabe N, Kyo S, Kondo S, Kanaya T, Wang Z, Maida Y, Takakura M, Nakamura M, Tanaka M, Inoue M. 2-5A antisense therapy directed against human telomerase RNA inhibits telomerase activity and induces apoptosis without telomere impairment in cervical cancer cells. Cancer Gene Ther. 2002;9:624-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 10. | Kubo T, Bakalova R, Ohba H, Fujii M. Antisense effects of DNA-peptide conjugates. Nucleic Acids Res Suppl. 2003;3:179-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 11. | Kosciolek BA, Kalantidis K, Tabler M, Rowley PT. Inhibition of telomerase activity in human cancer cells by RNA interference. Mol Cancer Ther. 2003;2:209-216. [PubMed] |
| 12. | Jansen B, Zangemeister-Wittke U. Antisense therapy for cancer--the time of truth. Lancet Oncol. 2002;3:672-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 118] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 13. | Mo Y, Gan Y, Song S, Johnston J, Xiao X, Wientjes MG, Au JL. Simultaneous targeting of telomeres and telomerase as a cancer therapeutic approach. Cancer Res. 2003;63:579-585. [PubMed] |
| 14. | Rezler EM, Bearss DJ, Hurley LH. Telomere inhibition and telomere disruption as processes for drug targeting. Annu Rev Pharmacol Toxicol. 2003;43:359-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 98] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 15. | Odago FO, Gerson SL. Telomerase inhibition and telomere erosion: a two-pronged strategy in cancer therapy. Trends Pharmacol Sci. 2003;24:328-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 16. | Ibanez de Caceres I, Frolova N, Varkonyi RJ, Dulaimi E, Meropol NJ, Broccoli D, Cairns P. Telomerase is frequently activated in tumors with microsatellite instability. Cancer Biol Ther. 2004;3:289-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 17. | Fukazawa T, Maeda Y, Sladek FM, Owen-Schaub LB. Development of a cancer-targeted tissue-specific promoter system. Cancer Res. 2004;64:363-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 18. | Kondo S, Tanaka Y, Kondo Y, Hitomi M, Barnett GH, Ishizaka Y, Liu J, Haqqi T, Nishiyama A, Villeponteau B. Antisense telomerase treatment: induction of two distinct pathways, apoptosis and differentiation. FASEB J. 1998;12:801-811. [PubMed] |
| 19. | Naka K, Yokozaki H, Yasui W, Tahara H, Tahara E, Tahara E. Effect of antisense human telomerase RNA transfection on the growth of human gastric cancer cell lines. Biochem Biophys Res Commun. 1999;255:753-758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Taggart AK, Teng SC, Zakian VA. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 290] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 21. | Wong SC, Yu H, Moochhala SM, So JB. Antisense telomerase induced cell growth inhibition, cell cycle arrest and telomerase activity down-regulation in gastric and colon cancer cells. Anticancer Res. 2003;23:465-469. [PubMed] |
| 22. | Shammas MA, Shmookler Reis RJ, Li C, Koley H, Hurley LH, Anderson KC, Munshi NC. Telomerase inhibition and cell growth arrest after telomestatin treatment in multiple myeloma. Clin Cancer Res. 2004;10:770-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 89] [Article Influence: 4.0] [Reference Citation Analysis (0)] |