Published online Apr 7, 2005. doi: 10.3748/wjg.v11.i13.1937
Revised: June 25, 2004
Accepted: July 17, 2004
Published online: April 7, 2005
AIM: To analyze the expression profiles of premalignant and/or preclinical lesions of gastric cancers.
METHODS: We analyzed the expression profiles of normal gastric pit, tubular adenoma and carcinoma in situ using microdissected cells from routine gastric biopsies. For the DNA microarray analysis of formalin-fixed samples, we developed a simple and reproducible RNA extraction and linear amplification procedure applying two polymerase-binding sites. The amplification procedure took only 8 h and yielded comparable DNA microarray data between formalin-fixed tissues and unfixed controls.
RESULTS: In comparison with normal pit, adenoma/carcinoma showed 504 up-regulated and 29 down-regulated genes at the expected false significance rate 0.15%. The differential expression between adenoma and carcinoma in situ was subtle: 50 and 22 genes were up-, and down-regulated in carcinomas at the expected false significance rate of 0.61%, respectively. Differentially expressed genes were grouped according to patterns of the sequential changes for the the ‘tendency analysis’ in the gastric mucosa-adenoma-carcinoma sequence.
CONCLUSION: Groups of genes are shown to reflect the sequential expression changes in the early carcinogenic steps of stomach cancer. It is suggested that molecular carcinogenic pathways could be analyzed using routinely processed biopsies.
-
Citation: Lee CH, Bang SH, Lee SK, Song KY, Lee IC. Gene expression profiling reveals sequential changes in gastric tubular adenoma and carcinoma
in situ . World J Gastroenterol 2005; 11(13): 1937-1945 - URL: https://www.wjgnet.com/1007-9327/full/v11/i13/1937.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i13.1937
Gastric cancer is one of the leading cancers worldwide[1]. Helicobacter pylori (H pylori) infection has been associated with gastric cancer[2], and the associated gastritis is regarded to be a major contributing factor in carcinogenesis[3,4]. H pylori-infection causes selective neutrophil infiltration to the proliferative zone of the gastric pits, which puts the actively regenerating cells under continuous mutagenic pressure and damage[5-7]. Acute foveolitis of the proliferative zone often induces extensive genomic damage in the proliferating cells which may be distinguished morphologically by the clear cellular changes, i.e., “the malgun cell changes”[6,7]. However, the premalignant lesions and/or carcinogenetic pathways have not been characterized[8].
Gastric cancers are diverse in the biological behavior as well as histogenesis. A considerable part of gastric cancers appears to develop from preexisting adenomas[9] while de novo carcinogenesis also exists[10]. They may be distinct not only in histogenesis but also in clinicopathological behaviors. To prevent and control gastric cancers, it would be important to figure out the carcinogenetic steps in both histopathological and molecular levels. The sequential expression changes from premalignant lesions to early stage cancers would provide insights into the carcinogenetic pathways and the detection of molecular targets for a specific treatment.
It has been a goal of oncogenomics to analyze the sequential expression changes at the premalignant and/or preclinical stage. Despite widespread application of DNA microarray analysis, it has remained to be a difficult goal to achieve because early lesions are often so small and subtle that they are only detected at the microscopic level convincingly, and consequently, are mostly available as formalin-fixed, paraffin-embedded samples as remnants of histopathological examination.
Once distinguished, the early lesions may be microdissected out from histological sections for the expression profiling analysis. Formalin-fixation provides excellent histological preparation for the detection of subtle premalignant lesions. For instance, the malgun cell change of gastric epithelium in H pylori gastritis may not be seen on frozen sections or other fixations[7]. Formalin-fixation and paraffin-embedding also confer the tissue stability for archival storage, and thus, has remained as the standard protocol for tissue preparation in pathology laboratories. However, formalin causes extensive base modifications of nucleic acids[11], which make it difficult to recover intact RNA and/or amplification necessary for the microarray analysis. Thus, a simple and reliable way of expression profiling of formalin-fixed tissue sections has been sought for as one of the bridges which would connect medicine with genomics.
Here, we present the expression profiles of normal gastric mucosa, adenoma, and carcinoma in situ using microdissected cells from formalin-fixed, paraffin-embedded sections. For the study, we have developed a simple RNA preparation/amplification procedure for the DNA microarray analysis applying two polymerase-binding sites. The amplification procedure including PCR and an in vitro transcription took only 8 h and provided comparable correlations with unfixed counterparts. Depending on the availability of microdissected cells, the procedure may be extended applying the second RNA polymerase. Using the procedure, more than 500 genes were detected to express differentially at the early stage of gastric carcinogenesis. They were analyzed in groups according to the patterns of sequential changes in the carcinogenetic pathway. Our data suggested that the screening for cancer related genes would be facilitated by the sequential expression changes at the early stage lesions using archival samples. The procedure and analysis were described in detail with pertinent information so that it may be reproduced readily in hospitals as well as research laboratories.
We used xenograft gastric cancer tissues for the development and fine adjustment of the RNA extraction/amplification method. Human gastric adenocarcinoma cell lines MKN45 and SNU484 were cultured in DMEM with 5% PBS. Cells were harvested with trypsinization when they reached 70% confluency. After being washed with PBS, ten million cells were resuspended in 0.3 mL PBS, and injected into nude mice subcutaneously. When the xenograft tumors reached 1 cm in diameter, mice were killed by cervical dislocation and tumors were harvested. MKN45 cells grew faster than SNU484 in culture and nude mice. Half of the tumors were frozen immediately in liquid nitrogen, and the rest were fixed in 10% buffered-formalin for 10 h at room temperature. Fixed tissue samples were processed for routine paraffin embedding.
Ten gastric biopsies having tubular adenomas and/or well-differentiated adenocarcinomas in situ were selected randomly from the surgical pathology file of Asan Medical Center, Seoul, Korea. Tubular adenomas were from eight male and two female patients ranging from 53 to 69 years old. Carcinomas were from 7 males and 3 females patients ranging from 45 to 74 years old. Controls were from 10 normal mucosa biopsies which were negative for H pylori infection. This study was approved by the Clinical Research Review Board of Asan Medical Center, Seoul, Korea.
Biopsies were fixed immediately in 10% buffered-formalin and processed routinely. After the histopathological diagnosis, additional 5 umol/L sections were taken from the paraffin blocks. For the sectioning and H and E staining, all the solutions were freshly made using DEPC-treated water, and the slides and instruments were autoclaved. Cells were microdissected using an AutoPix laser capture microscope system (Arcturus, Mountain View, CA.).
Total RNA was extracted twice from freshly frozen xenograft tissues using TRIzol reagents (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. For the RNA extraction from formalin-fixed tissues, deparaffinized sections were removed from the slides by applying 200 proteinase K buffer [2% SDS, 10 mmol/L Tris-HCl (pH 8.0), 0.1 mmol/L EDTA]. Samples were transferred into a microcentrifuge tube and incubated at 70 °C for 1 h to relieve the formalin-induced modifications. Then, 3 μL proteinase K (30 μg/μL, Intron biotechnology, Songnam, Korea) was added, and incubated again at 55 °C for 1 h. RNAs were extracted with TRIzol similarly. The extracted RNAs were precipitated in isopropanol with 5 μg linear acrylamide (Ambion, Austin, TX), and the RNA pellets were resuspended in 10 μL nuclease-free water (Ambion). The quality of extracted RNAs was checked using denaturing agarose gels. The amount of extracted RNA samples and/or amplified RNA was measured using a RiboGreen RNA quantitation kit (Molecular probes, Eugene, Oregon) according to the manufacturer’s protocol. Each measurement was duplicated, and the average values were taken.
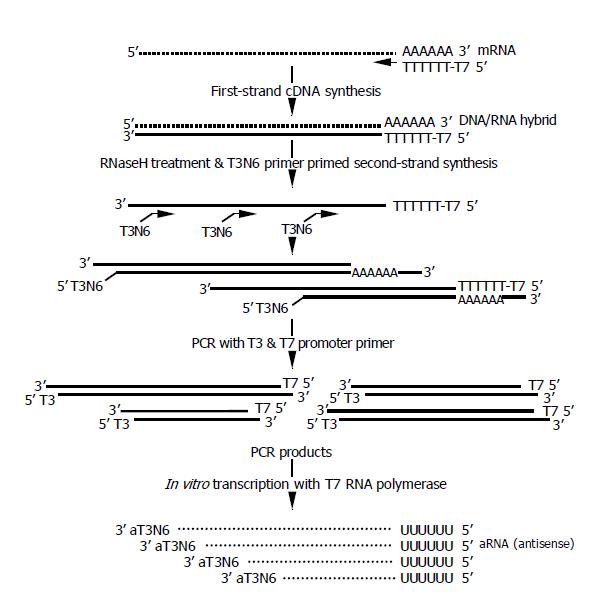
For the first strand synthesis, 100 pmoL of T7dT primers (100 pmol/μL, 5’- AAACGACGGCCAGTGAATTGTA-ATACGACTCACTATAGGCGCTTTTTTTTTTTTTTT-TTTTTTTTT-3’, Bioneer, Daejon, Korea) was added to the 10 μL of total RNA, and incubated at 70 °C for 10 min. After primers were let to anneal to RNA templates by incubating on ice for 10 min, 4 μL 5X first-strand buffer, 0.5 μL RNase inhibitor (40 U/μL, Promega, Madison, WI), 2 μL 0.1 mol/L DTT, 1 μL dNTPmix (10 mmol/L each, Roche, Mannheim, Germany), and 2 μL SuperScript IITM reverse transcriptase (Invtirogen, Carlsbad, CA) were added. The mixture was incubated at 42 °C for 2 h.
For the second strand synthesis, 1 μL RNAse H (2 U/μL, Invitrogen, Carlsbad, CA) was added to the mixture, and incubated at 37 °C for 15 min, and at 95 °C for 2 min. Then, 1 μL random T3N6 primers (100 pmoles/μL, 5’-GCGCGAAATTAACCCTCACTAAAGGGAGANNNNNN-3’) were added, incubated at 95 °C for 2 min, and placed on ice for 10 min. Then, 20 μL 5X second-strand buffer (Invtirogen, Carlsbad, CA), 2 μL dNTPmix (10 mm each), nuclease-free water 53.5 μL, 2.5 μL E. coli DNA polymerase I (10 u/μL, Invitrogen, Carlsbad, CA) were added and incubated at 16 °C for 2 h. The synthesized double-stranded DNA was purified using a MinEluteTM PCR purification kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. To retrieve DNA efficiently, samples were eluted twice with 42 μL eluting solution.
Then the double stranded DNA was applied to PCR amplification. To the elution, 10 μL 10X Advantage 2 PCR buffer (Clontech), 1 μL T7 promoter primers (100 pmol/μL, 5’-CGGCCAGTGAATTGTAATACGACTCACT-ATAGGCG-3’), 1 μL T3 promoter primers (100 pmol/μL, 5’-GCGCGAAATTAACCCTCACTAAAGGGAGAGGG-3’), 2 μL 10 mmol/L dNTP mix, and 2 μL advantage 2 polymerase mix (Clontech) were added. PCR reaction was done in a Gene-Amp PCR 9600 system (PE Biosystems, Foster City, CA) for 1 min at 95 °C, 20 cycles for 30 s at 95 °C for 40 s at 65 °C for 5 min at 68 °C, for 7 min at 68 °C. PCR products were purified using a MinEluteTM PCR purification kit (Qiagen, Valencia, CA), and were eluted using 10 μL nuclease-free water twice.
Following the PCR amplification, aRNA synthesis (in vitro transcription) was performed using an AmpliScribeTM T7 high yield transcription kit (Epicenter, Madison, WI) at 37 °C for 5 h in 40 μL of reaction volume. Synthesized aRNA was purified using a RNeasy Mini kit (Qiagen).
For cDNA microarray analysis, stomach cancer-specific 14K cDNA microarray chips were applied[12]. The probe labeling and hybridization were done using the amine-modified random primer aminoallyl method[13]. Probes were synthesized from 10 μg unamplified total RNA or 30 μg aRNA using 2 μg amine-modified random primer (5’-C6dTNNNNN-3’, SIGMA Genosys, The Woodlands, TX). For the xenograft study, SNU484 and MKN45 tumor samples were labeled with Cy3 and Cy5, respectively. For the microdissection study, either adenoma or carcinoma was labeled with Cy3 and hybridized against the same normal control labeled with Cy5. The labeled probes were mixed, and the volume was adjusted to 500 μL by adding nuclease-free water. The final volume was adjusted to 17 μL by centrifugation (10000 g) in the Microcon YM-30 (Millipore, Bedford, MA).
For the hybridization, 1 μL poly A (8 mg /mL, Roche, Mannheim, Germany), 1 μL Cot-1 DNA (10 mg/mL, Invitrogen, Carlsbad, CA), 1 μL yeast tRNA (4 mg/mL, Invitrogen, Carlsbad, CA) were added to the labeled probe, and denatured at 100 °C for 2 min, and cooled on ice. The probe was mixed with 20 μL 2×formamide hybridization buffer [50% formamide, 10×SSC, 0.2% SDS], and applied to the DNA microarray. A glass cover slip was applied, and the microarray was put in the hybridization cassette (TeleChem International, Sunnyvale, CA). After being incubated overnight in a 42 °C water-bath, microarrays were washed with the first [2×SSC, 0.1% SDS] and second wash solutions [0.5×SSC, 0.01% SDS] for 5 min, respectively. Remaining water was removed from the slide by centrifugation at 800 r/min for 2 min.
The arrays were scanned with a GenePix 4000B scanner (Axon, Foster City, CA) at 10 µm resolution. The PMT voltage settings were varied to obtain the maximum signal intensities with <1% probe saturation. The resulting images were analyzed using the ImaGeneTM 4.0 (BioDiscovery, Los Angeles, CA) software. Spots having a signal-to-noise ratio over 1.4 were screened and normalized for the analysis. Pearson correlation coefficients of the global and differentially expressed genes were calculated using the SPSS software (SPSS Inc. Chicago, IL).
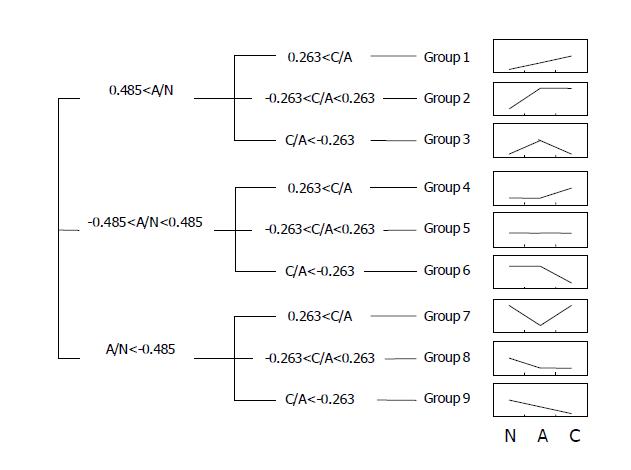
SAM analysis was done to detect differentially expressed genes in microdissected adenomas and carcinomas[14]. The selected genes were divided into nine groups to show the patterns of sequential changes among the normal pits, adenoma, and carcinoma. First, genes were divided into three categories arbitrarily according to the expression changes in adenomas compared to the control: up-regulated (>1.4 times, log ratio >0.485), down-regulated (<1.4 times), and ‘unchanged’. Then, each category was further divided into three groups according to the expression changes in carcinomas compared to the control: up-regulated (>1.2 times, log ratio >0.263), down-regulated (<1.2 times), and ‘unchanged’.
Both MKN45 and SNU484 xenograft tumors consisted of solid cell clusters, being reminiscent of poorly differentiated gastric adenocarcinomas (data not shown). The expression profiles were analyzed using unamplified samples, and the data were used as a control for the development and fine adjustment of the amplification conditions in the subsequent experiments using formalin-fixed counterparts. The microarray data including those of microdissected samples were deposited at the GEO (http://www.ncbi.nlm.nih.gov/geo/) (accession numbers GSM20670-5).
The expression profiling of MKN45 and SNU484 xenograft tumors showed that they had quite distinct expression patterns despite the histopathological similarity. The up-regulated genes in SNU484 in comparison with MKN45 included aldehyde dehydrogenase 1 family member A1 (ALDH1A1), thymosin beta 4 (TMSB4X), activated leukocyte cell adhesion molecule (ALCAM), collagen type XI alpha 1 (COL11A1), and plectin 1 (PLEC1), etc. The up-regulated genes in MKN45 tumor included EBNA2 co-activator (p100), regenerating gene type-4 (REG-IV), S100 calcium binding protein A4 (S100A4), replication initiation region protein (60 ku) (RIP60), and carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6), etc. Considering that MKN45 cells grew much faster than SNU484 cells in vitro and ex vivo, some of the differentially expressed genes might be related to cell growth and/or aggressiveness. For instance, S100A4 protein up-regulation was shown to associate with metastasis and poor prognosis of stomach cancer and others[15].
It was reported that sample heating relieved the extensive base modifications of nucleic acids induced by formalin-fixation[11]. To make the procedure as simple as possible, we used only heating/proteinase K treatment to observe how efficient and reproducible the RNA extraction was. Samples were heated at 70 °C for 1 h before the proteinase K treatment for the RNA extraction. RNAs were extracted from the fixed tissue sections of SNU484 and MKN45 xenograft tumors using the protocol. The extracted total RNAs were estimated to be 18.3 and 24.1 pg per fixed cell of MKN45 and SNU484 tumors, respectively (Table 1). They corresponded to 63.1 and 77.7% of the total RNAs of MKN45 (29 pg) and SNU484 (31 pg) cells in culture, suggesting that the extraction efficiency from formalin-fixed cells was comparable to that from fresh counterparts.
| Sample | AmplificationMethod | Starting materials | Amplified RNA(ng) | 3Amplification Fold (X) | |
| Total RNA (ng) (1mRNA) | 2Cell number | ||||
| Fresh MKN45 | 690 | 152.2 | 761 X | ||
| 4T7 IVT only | 20 (0.2) | ||||
| SNU484 | 650 | 213.2 | 1066 X | ||
| Fixed MKN45 | 830 | 121.6 | 608 X | ||
| T7 IVT only | 20 (0.2) | ||||
| SNU484 | 1090 | 128.6 | 643 X | ||
| MKN45 | 830 | 42120 | 210600 X | ||
| 5PCR-T7 IVT | 20 (0.2) | ||||
| SNU484 | 1090 | 45016 | 225080 X | ||
The amplification procedure is depicted in Figure 1. The entire procedure of RNA extraction and one-round amplification took only 8 h. The first strand cDNA was synthesized using T7dT primers as described previously[16,17]. Then, the second strand were synthesized using T3N6 primers, and 20 cycles of PCR amplification was done using the T3 and T7 promoter primers as described in the Materials and Methods. The resulting PCR products were used for the in vitro transcription using either T7 or T3 polymerase, which yielded similar amplification rates (data not shown). If the amplified products were not enough for the microarray analysis, in vitro transcription might be repeated using either T3 or T7 polymerase, whichever was unused in the first round.
The amplification results from two xenograft tumors are summarized in Table 1. To see whether RNAs from formalin-fixed samples were adequate for the amplification, we first checked the amplification only in vitro transcription using T7 polymerase. The amounts of aRNAs of fixed MKN45 and SNU484 tumors were increased 6.43 and 6.08 times, respectively. Provided the mRNA amount was 1% of the total RNAs, the amplification rates were estimated to be 643 and 608 times, respectively. For the fresh samples, the amplification rates after the in vitro transcription were estimated to be 761 and 1 066 times for MKN45 and SNU484 tumors, respectively, showing that the amplification efficiencies of fixed samples were comparable to those of fresh counterparts. When fixed MKN45 and SNU484 tumors were processed for a complete round of amplification, the amplification rates were 210600 and 225080 times, respectively. Since 20-40 μg of aRNA was enough for a DNA microarray depending on the hybridization methods, it was estimated that less than 1000 cells were required for a DNA microarray analysis.
Amplified aRNAs were hybridized on 14K cDNA microarrays, and the data were compared with those of the unamplified fresh tissue controls. Upon the filtering, 13706 and 13407 spots were left for the aRNA and controls, respectively. The correlation coefficient of global gene expression was 0.718. The correlation coefficient was increased to 0.858, when 500 genes differentially expressed between SNU484 and MKN45 more than twice were compared.
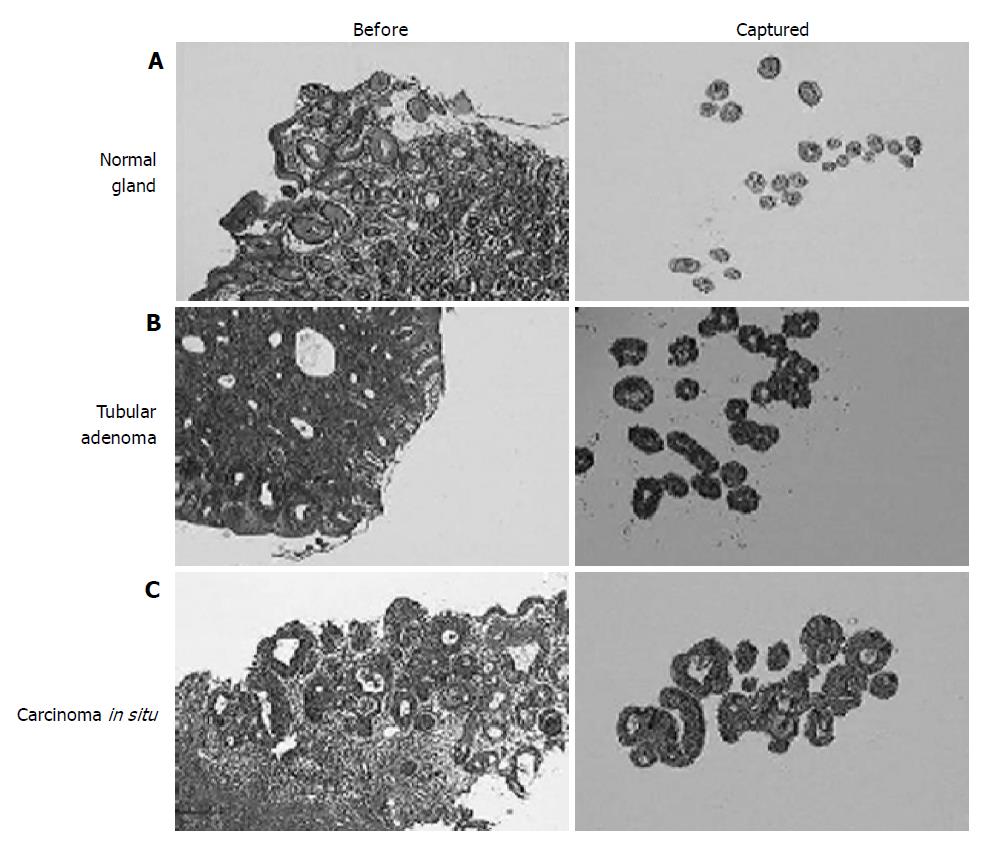
We, then, analyzed the expression profiles of normal gastric epithelium, adenoma, and adenocarcinoma in situ using microdissected cells from ‘routine’ gastric biopsies. For the sequential analysis of early lesions only the carcinomas without stromal invasion were included. For the normal control, pit epithelia were taken only from the proliferative zone in order to minimize the ‘contamination’ by genes related to nonspecific cell proliferation in adenomas and carcinomas (Figure 2A). To avoid cells with DNA damage, any epithelial cells showing the malgun cell change were excluded[7]. Adenomas had glands which were rather regular in size and orientation, which were much more than the normal glandular distribution (Figure 2B). Epithelial cells were uniformly columnar and well oriented. Nuclei were hyperchromatic with a high nucleus/cytoplasm ratio. Carcinomas consisted of glands which were mildly irregular in size and arrangement in comparison with those of adenomas (Figure 2C). Cells were not particularly different from adenoma cells in size, but nuclei were ovoid and nucleoli were more prominent.
For the experiment, a total of 10000 cells from 10 adenomas and/or carcinomas and 20000 normal control cells were microdissected and pooled, respectively. After the amplification, 135.2, 77.3, and 77.9 ug of aRNAs were obtained from the normal pit, adenoma, and carcinoma, respectively. Microarray hybridization was duplicated for adenomas and carcinomas, respectively, using the same control of normal pit epithelium.
The DNA microarray data were analyzed using the SAM method[14]. At the expected false significance rate of 0.15%, 504 up-regulated and 29 down-regulated genes were detected in adenoma and carcinoma (Figure 3A). Genes with differential expressions are summarized in Tables 2 and 3. The difference of expression between adenoma and carcinoma was subtle. Fifty up-regulated and 22 down-regulated genes were detected in carcinomas compared to adenomas at the expected false significance rate of 0.61% (Figure 3B).
| Symbol | Annotation | GenBank |
| NUP153 | Nucleoporin 153ku | NM_005124 |
| CENPA | Centromere protein A (17 ku) | NM_001809 |
| RPL23 | Ribosomal protein L23 | NM_000978 |
| LMAN2 | Lectin, mannose-binding 2 | NM_006816 |
| KIAA0469 | KIAA0469 gene product | NM_014851 |
| POLR2C | Polymerase (RNA) II (DNA directed) polypeptide C, 33 ku | NM_002694 |
| G2AN | Alpha glucosidase II alpha subunit | NM_014610 |
| KIAA0007 | KIAA0007 protein | D26488 |
| EPHB4 | EphB4 | NM_004444 |
| YWHAQ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | NM_006826 |
| DHX9 | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | NM_001357 |
| K-ALPHA-1 | Tubulin, alpha, ubiquitous | NM_006082 |
| CPR2 | Cell cycle progression 2 protein | NM_030900 |
| PICALM | Phosphatidylinositol binding clathrin assembly protein | NM_007166 |
| CXCL5 | Chemokine (C-X-C motif) ligand 5 | NM_002994 |
| MGC3103 | KUGI seq X - hypothetical protein MGC3103 | NM_024036 |
| CHUK | Conserved helix-loop-helix ubiquitous kinase | NM_001278 |
| FLJ20297 | Hypothetical protein FLJ20297 | NM_017751 |
| RPL4 | Ribosomal protein L4 | NM_000968 |
| TFPI | Tissue factor pathway inhibitor (lipoprotein-associated coagulationinhibitor) | NM_006287 |
| CD9 | CD9 antigen (p24): tetraspanin superfamily | NM_001769 |
| ZFP276 | Zinc finger protein 276 | NM_152287 |
| HYOU1 | Hypoxia up-regulated 1 | NM_006389 |
| HTATIP | HIV-1 Tat interactive protein, 60 ku | NM_006388 |
| TMPO | Thymopoietin | NM_003276 |
| FLJ10521 | Hypothetical protein FLJ10521 | NM_018125 |
| GBAS | Glioblastoma amplified sequence | NM_001483 |
| LOC51142 | 16.7 ku protein | NM_016139 |
| LGTN | Ligatin | NM_006893 |
| NEDD5 | Neural precursor cell expressed, developmentally down-regulated 5 | NM_004404 |
| PAFAH1B2 | Platelet-activating factor acetylhydrolase, isoform Ib, beta subunit 30 ku | NM_002572 |
| RPIA | Ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) | NM_144563 |
| ARHGEF9 | Cdc42 guanine nucleotide exchange | NM_015185 |
| RNPS1 | RNA binding protein S1, serine-rich domain | NM_006711 |
| RPL27 | Ribosomal protein L27 | NM_000988 |
| RAMP | RA-regulated nuclear matrix-associated protein | NM_016448 |
| PRKAG1 | Protein kinase, AMP-activated, gamma 1 non-catalytic subunit | NM_002733 |
| HMGB1 | High-mobility group box 1 | NM_002128 |
| TOP1MT | Mitochondrial topoisomerase I | NM_052963 |
| HNRPA0 | Heterogeneous nuclear ribonucleoproteinA0 | NM_006805 |
| TRA1 | Tumor rejection antigen (gp96) 1 | NM_003299 |
| BAP1 | BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) | NM_004656 |
| SMARCD2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 | NM_003077 |
| FLJ11749 | Hypothetical protein FLJ11749 | NM_024591 |
| RPL37 | Ribosomal protein L37 | NM_000997 |
| RPS17 | Ribosomal protein S17 | NM_001021 |
| HNRPM | Heterogeneous nuclear ribonucleoprotein M | NM_005968 |
| AP3B1 | Adaptor-related protein complex 3, beta 1 subunit | NM_003664 |
| CRR9 | Cisplatin resistance related protein CRR9p | NM_030782 |
| KIAA0152 | KIAA0152 gene product | NM_014730 |
| TFF2 | Trefoil factor 2 (spasmolytic protein 1) | NM_005423 |
| GALIG | Galectin-3 internal gene | NM_194327 |
| TSPAN-1 | Tetraspan 1 | NM_005727 |
| MUC5AC | Mucin 5, subtypes A and C, tracheobronchial/gastric | AJ001403 |
| REG-IV | Regenerating islet-derived family, member 4 | NM_032044 |
| LOC376711 | Similar to calpain 8 | XM_352343 |
| SYTL2 | Synaptotagmin-like2 | NM_032943 |
| MT2A | Metallothionein2A | NM_005953 |
| RPL13A | Ribosomal protein L13a | NM_012423 |
| DPCR1 | Diffuse panbronchiolitis critical region | NM_080870 |
| IGL@ | Immunoglobulin lambda locus | BM918733 |
| TFF1 | Trefoil factor 1 (breast cancer, estrogen-inducible sequence expressed in) | NM_003225 |
| GUK1 | Guanylate kinase 1 | NM_000858 |
| SLC25A19 | Solute carrier family 25 (mitochondrial deoxynucleotide carrier), member 19 | NM_021734 |
| LGALS3 | Lectin, galactoside-binding, soluble, 3 | NM_002306 |
| PSCA | Prostate stem cell antigen | NM_005672 |
| FLJ38641 | FLJ38641 | AL358512 |
| TSPAN-3 | Tetraspan 3 | NM_005724 |
| KCNE3 | Potassium voltage-gated channel, Isk-related family, member 3 | NM_005472 |
| GDDR | Down-regulated in gastric cancerGDDR | NM_182536 |
| SERPINH1 | Serine (or cysteine) proteinase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) | NM_001235 |
| ZNF265 | Zinc finger protein 265 | NM_005455 |
| CYP2S1 | Cytochrome P450, family 2, subfamily S, polypeptide 1 | NM_030622 |
| AGR2 | Anterior gradient 2 (Xenepus laevis) homolog | NM_006408 |
To analyze the genes according to the patterns of sequential expression changes, we further divided the genes into nine groups as described in the Materials and Methods (Figure 4). Groups 1, 2, and 3 included genes that were up-regulated in adenomas, and kept on being up-regulated, maintained, and down-regulated in carcinomas (47, 178, and 117 genes), respectively. Groups 4, 5, and 6 included genes with minimal to mild variations in adenomas, which were then up-regulated, maintained, and down-regulated in carcinomas (57, 73, and 2 genes), respectively. Groups 7, 8, and 9 included genes which were down-regulated in adenomas, and then, up-regulated, maintained, and kept on being down-regulated in carcinomas (28, 10, and 9 genes), respectively. It was expected that such a ‘tendency analysis’ at multiple points of carcinogenetic steps would help recognize groups of genes with a distinct biological significance at the early stage of gastric carcinogenesis. As discussed later, many genes associated with cancers were included in the groups that were expected to implicate in the carcinogenesis significantly. Many novel genes with unknown functions were also included in each group.
We analyzed the expression profiles of early lesions of gastric cancer using formalin fixed biopsies. Our goal was to develop a simple and practical procedure for the RNA extraction and amplification that may be applied to the reproducible analysis of formalin-fixed samples in hospitals and research laboratories. It has been suggested that the RNA extracted from formalin-fixed tissues using various methods could be used for quantitative analysis in many fields of biological research[18-25]. Our data suggested that the simple procedure could improve not only the extraction efficiency but also the quality of RNA that were good enough for the DNA microarray analysis.
Our strategy for the amplification was to introduce two RNA polymerase binding sites that might be used for both PCR and in vitro transcription (s). We applied T7dT primers and random T3N6 primers to the first and second strand DNA synthesis, respectively. The T3N6 primers yielded a similar reproducibility but a better efficiency in comparison with T3N9 primers, which were shown to reproduce similar results after repeated rounds of amplifications[17] (data now shown). Our ‘2 binding sites’ strategy reduced the unnecessary preparation time for each step so that the entire procedure of PCR and one round in vitro transcription took only 8 h. When it was necessary, the second round of in vitro transcription might be added using either T3 or T7 RNA polymerase, whichever was not used in the first round. Alternatively, two consecutive rounds of in vitro transcriptions might be done omitting the PCR step, although it would be more time-consuming. In any case, the flexibility is of a critical advantage when the availability of cells is limited. Because the bias induced by the random primer hybridization would have already been reflected in the PCR products, the second round amplification was not expected to introduce significant errors.
The PCR was applied to the linear amplification for DNA microarray analysis[26,27]. Iscove et al[26] limited the elongation time so that only extreme 3’ ends of similar length were amplified. Aoyagi et al[27] did the in vitro transcription first, and then, applied adaptors to the cDNAs for the PCR amplification. In contrast, our method was simple and did not require any additional procedure for the PCR. We allowed sufficient elongation time to assure a complete cycle for each cDNA. PCR amplification was also applied to the simultaneous amplification of multiple genes[25].
Formalin-fixed tissues produced comparable data with those of the unfixed control (global correlation coefficient 0.718). Interestingly, the correlation coefficients increased considerably when only differentially expressed genes were analyzed. The increased correlation may suggest that the amplification of abundant genes is relatively privileged. Anyway, it is an encouraging finding for the practical application, because the aim of most DNA microarray screening is to detect the differentially expressed genes rather than global gene profiling.
Our data suggested that the subtle differential expression between adenoma and carcinoma in situ could be detected convincingly. DNA microarray analysis was based on competitive hybridization, and so far, most studies have been designed to compare the expression profiles of two distinct lesions that may or may not be directly related. Our approach of multi-point comparison may provide a unique opportunity for the “tendency analysis”, which could be helpful for the screening of biologically significant genes in the carcinogenesis and progression of diseases.
Groups 1, 2, and 3 included genes that were up-regulated in adenomas, and kept on being up-regulated, maintained, and down-regulated in carcinomas. They included many genes that were up-regulated in cancers and implicated in the carcinogenesis, suggesting that our data were quite reproducible. Group 1 included many genes which were implicated in carcinogenesis and/or up-regulated in cancers: junction plakoglobin (gamma catenin: JUP), squamous cell carcinoma antigens recognized by T cell 3 (SART3), DEAH (Asp-Glu-Ala-His) box polypeptide 9 (DHX9), mucin 4 (MUC4), ribosomal protein L15 (RPL15), microphthalmia-associated transcription factors(MITF), and fusion (involved in t (12;16) in malignant liposarcoma) (FUS). JUP and MITF have been implicated in the Wnt pathway, which is one of the main carcinogenic pathways[28,29].
Groups 2 and 3 included A-Raf (v-raf murine sarcoma 3611 viral oncogene homolog 1: ARAF1), v-akt murine thymoma viral oncogene homolog 2 (AKT2), v-jun sarcoma virus 17 oncogene homolog (JUN), glioblastoma amplified sequence (GBAS), tumor rejection antigen 1 (TRA1), tumor protein D52-like 2 (TPD52L2), hypoxia up-regulated 1 (HYOU1), heparan sulfate proteoglycan 2 (HSPG2), polo-like kinase (PLK), high-mobility group box 1(HMGB1), and tumor protein, translationally-controlled 1 (TPT1). In addition, nuclear proteins such as nucleolin, nucleolar protein family A, member 1 (NOLA1), nucleoporin 153 ku (NUP153), nucleoporin (Nup37), and lamin B receptor (LBR) were also included. The up-regulation of nucleolar proteins was compatible with the continuous enlargement of nucleoli in adenoma and carcinomas.
In group 4, genes up-regulated in carcinoma but not in adenoma, villin 2 (VIL2) and S100A4 were included. Villin 2 plays a key role in cell surface structure adhesion, migration, and organization, and has been reported to be associated with invasiveness of esophageal carcinoma[30]. S100A4 was also up-regulated in MKN45 cells in comparison with SNU484 cells, and was shown to associate with metastasis and poor prognosis of stomach cancers and others[15]. Nucleolin was also included in this group. Group 6 consisted of two genes that were down regulated in carcinoma: progesterone receptor membrane component 2 (PGRMC2) and acyl-coenzyme A dehydrogenase, C-2 to C-3 short chain (ACADS).
Groups 7, 8, and 9 included down-regulated genes in gastric cancers. Group 7 included mucin 5 subtypes A and C, tracheobronchial/gastric (MUC5AC) and trefoil factor 1 (TFF1), and group 8 included fatty acid binding protein 1 (FABP1). TFF1 has been reported to be down-regulated in most gastric cancers[31]. It should be noted that FABP1 is one of the tamoxifen-target proteins, the block of which might be related to the anti-cancer effect [32]. Group 9 included trefoil factor 2 (TFF2) which was down-regulated in gastric cancer (GDDR). GDDR was reported to be a down-regulated gene in stomach cancer[33]. It is a transmembrane protein homologous to carbonic anhydrase-like CA11[34].
In conclusion, differential expressions between lesions reflecting subtle histopathological changes may be detected using microdissected cells from formalin-fixed tissues. The sequential expression analysis at multiple points in a pathogenic pathway facilitates the detection of biologically relevant genes in the development and progression of diseases. The protocol may be applied to the search for various disease-related genes using archival tissue samples.
| 1. | Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer. 1999;80:827-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 2. | Plummer M, Franceschi S, Muñoz N. Epidemiology of gastric cancer. IARC Sci Publ. 2004;157:311-326. [PubMed] |
| 3. | Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3221] [Cited by in RCA: 3624] [Article Influence: 120.8] [Reference Citation Analysis (6)] |
| 4. | Genta RM. The gastritis connection: prevention and early detection of gastric neoplasms. J Clin Gastroenterol. 2003;36:S44-S49; discussion S61-S62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Yu E, Lee HK, Kim HR, Lee MS, Lee I. Acute inflammation of the proliferative zone of gastric mucosa in Helicobacter pylori gastritis. Pathol Res Pract. 1999;195:689-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 6. | Lee H, Jang J, Kim Y, Ahn S, Gong M, Choi E, Lee I. "Malgun" (clear) cell change of gastric epithelium in chronic Helicobacter pylori gastritis. Pathol Res Pract. 2000;196:541-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 7. | Jang J, Lee S, Jung Y, Song K, Fukumoto M, Gould VE, Lee I. Malgun (clear) cell change in Helicobacter pylori gastritis reflects epithelial genomic damage and repair. Am J Pathol. 2003;162:1203-1211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 8. | Misdraji J, Lauwers GY. Gastric epithelial dysplasia. Semin Diagn Pathol. 2002;19:20-30. [PubMed] |
| 9. | Kase S, Osaki M, Honjo S, Adachi H, Ito H. Tubular adenoma and intramucosal intestinal-type adenocarcinoma of the stomach: what are the pathobiological differences? Gastric Cancer. 2003;6:71-79. [PubMed] |
| 10. | Tahara E. Genetic pathways of two types of gastric cancer. IARC Sci Publ. 2004;157:327-349. [PubMed] |
| 11. | Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27:4436-4443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 475] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 12. | Kim B, Bang S, Lee S, Kim S, Jung Y, Lee C, Choi K, Lee SG, Lee K, Lee Y. Expression profiling and subtype-specific expression of stomach cancer. Cancer Res. 2003;63:8248-8255. [PubMed] |
| 13. | Xiang CC, Kozhich OA, Chen M, Inman JM, Phan QN, Chen Y, Brownstein MJ. Amine-modified random primers to label probes for DNA microarrays. Nat Biotechnol. 2002;20:738-742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 14. | Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116-5121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9125] [Cited by in RCA: 9163] [Article Influence: 366.5] [Reference Citation Analysis (0)] |
| 15. | Yonemura Y, Endou Y, Kimura K, Fushida S, Bandou E, Taniguchi K, Kinoshita K, Ninomiya I, Sugiyama K, Heizmann CW. Inverse expression of S100A4 and E-cadherin is associated with metastatic potential in gastric cancer. Clin Cancer Res. 2000;6:4234-4242. [PubMed] |
| 16. | Eberwine J. Single-cell molecular biology. Nat Neurosci. 2001;4 Suppl:1155-1156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 17. | Xiang CC, Chen M, Kozhich OA, Phan QN, Inman JM, Chen Y, Brownstein MJ. Probe generation directly from small numbers of cells for DNA microarray studies. Biotechniques. 2003;34:386-388, 390, 392-393. [PubMed] |
| 18. | Godfrey TE, Kim SH, Chavira M, Ruff DW, Warren RS, Gray JW, Jensen RH. Quantitative mRNA expression analysis from formalin-fixed, paraffin-embedded tissues using 5' nuclease quantitative reverse transcription-polymerase chain reaction. J Mol Diagn. 2000;2:84-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 244] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 19. | Specht K, Richter T, Müller U, Walch A, Werner M, Höfler H. Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol. 2001;158:419-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 365] [Article Influence: 14.6] [Reference Citation Analysis (6)] |
| 20. | Van Deerlin VM, Gill LH, Nelson PT. Optimizing gene expression analysis in archival brain tissue. Neurochem Res. 2002;27:993-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 21. | Cohen CD, Gröne HJ, Gröne EF, Nelson PJ, Schlöndorff D, Kretzler M. Laser microdissection and gene expression analysis on formaldehyde-fixed archival tissue. Kidney Int. 2002;61:125-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 79] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 22. | Karsten SL, Van Deerlin VM, Sabatti C, Gill LH, Geschwind DH. An evaluation of tyramide signal amplification and archived fixed and frozen tissue in microarray gene expression analysis. Nucleic Acids Res. 2002;30:E4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 106] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 23. | Körbler T, Grsković M, Dominis M, Antica M. A simple method for RNA isolation from formalin-fixed and paraffin-embedded lymphatic tissues. Exp Mol Pathol. 2003;74:336-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 52] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 24. | Abrahamsen HN, Steiniche T, Nexo E, Hamilton-Dutoit SJ, Sorensen BS. Towards quantitative mRNA analysis in paraffin-embedded tissues using real-time reverse transcriptase-polymerase chain reaction: a methodological study on lymph nodes from melanoma patients. J Mol Diagn. 2003;5:34-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 137] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 25. | Cronin M, Pho M, Dutta D, Stephans JC, Shak S, Kiefer MC, Esteban JM, Baker JB. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol. 2004;164:35-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 420] [Article Influence: 19.1] [Reference Citation Analysis (7)] |
| 26. | Iscove NN, Barbara M, Gu M, Gibson M, Modi C, Winegarden N. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat Biotechnol. 2002;20:940-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 212] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 27. | Aoyagi K, Tatsuta T, Nishigaki M, Akimoto S, Tanabe C, Omoto Y, Hayashi Si, Sakamoto H, Sakamoto M, Yoshida T. A faithful method for PCR-mediated global mRNA amplification and its integration into microarray analysis on laser-captured cells. Biochem Biophys Res Commun. 2003;300:915-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 71] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 28. | Kolligs FT, Kolligs B, Hajra KM, Hu G, Tani M, Cho KR, Fearon ER. gamma-catenin is regulated by the APC tumor suppressor and its oncogenic activity is distinct from that of beta-catenin. Genes Dev. 2000;14:1319-1331. [PubMed] |
| 29. | Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1-24. [PubMed] |
| 30. | Shen ZY, Xu LY, Chen MH, Li EM, Li JT, Wu XY, Zeng Y. Upregulated expression of Ezrin and invasive phenotype in malignantly transformed esophageal epithelial cells. World J Gastroenterol. 2003;9:1182-1186. [PubMed] |
| 31. | Beckler AD, Roche JK, Harper JC, Petroni G, Frierson HF, Moskaluk CA, El-Rifai W, Powell SM. Decreased abundance of trefoil factor 1 transcript in the majority of gastric carcinomas. Cancer. 2003;98:2184-2191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 32] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 32. | Mésange F, Sebbar M, Capdevielle J, Guillemot JC, Ferrara P, Bayard F, Poirot M, Faye JC. Identification of two tamoxifen target proteins by photolabeling with 4-(2-morpholinoethoxy)benzophenone. Bioconjug Chem. 2002;13:766-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 33. | Du JJ, Dou KF, Peng SY, Wang WZ, Wang ZH, Xiao HS, Guan WX, Liu YB, Gao ZQ. Down-regulated full-length novel gene GDDR and its effect on gastric cancer. Zhonghua YiXue ZaZhi. 2003;83:1166-1168. [PubMed] |
| 34. | Shiozaki K, Nakamori S, Tsujie M, Okami J, Yamamoto H, Nagano H, Dono K, Umeshita K, Sakon M, Furukawa H. Human stomach-specific gene, CA11, is down-regulated in gastric cancer. Int J Oncol. 2001;19:701-707. [PubMed] |