Published online Jan 7, 2005. doi: 10.3748/wjg.v11.i1.36
Revised: April 15, 2004
Accepted: May 13, 2004
Published online: January 7, 2005
AIM: To design a novel method to rapidly detect the quantitative alteration of mtRNA in patients with tumors.
METHODS: Oligo 6.22 and Primer Premier 5.0 bio-soft were used to design 15 pairs of primers of mtRNA cDNA probes in light of the functional and structural property of mtDNA, and then RT-PCR amplification was used to produce 15 probes of mtRNA from one normal gastric mucosal tissue. Total RNA extracted from 9 gastric cancers and corresponding normal gastric mucosal tissues was reverse transcribed into cDNA labeled with fluorescein. The spotted mtDNA microarrays were made and hybridized. Finally, the microarrays were scanned with a GeneTACTM laser scanner to get the hybridized results. Northern blot was used to confirm the microarray results.
RESULTS: The hybridized spots were distinct with clear and consistent backgrounds. After data was standardized according to the housekeeping genes, the results showed that the expression levels of some mitochondrial genes in gastric carcinoma were different from those in the corresponding non-cancerous regions.
CONCLUSION: The mtDNA expression microarray can rapidly, massively and exactly detect the quantity of mtRNA in tissues and cells. In addition, the whole expressive information of mtRNA from a tumor patient on just one slide can be obtained using this method, providing an effective method to investigate the relationship between mtDNA expression and tumorigenesis.
- Citation: Han CB, Mao XY, Xin Y, Wang SC, Ma JM, Zhao YJ. Quantitative analysis of tumor mitochondrial RNA using microarray. World J Gastroenterol 2005; 11(1): 36-40
- URL: https://www.wjgnet.com/1007-9327/full/v11/i1/36.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i1.36
Tumor mitochondria often differ very significantly from their normal counterparts. Alterations in organelles accompanying neoplastic transformation of cells are reflected in mitochondrial morphology, enzymatic composition and cellular content[1,2]. Mitochondrial DNA (mtDNA) is a 16569 bp double-stranded, closed circular molecule, which codes for a small (12S) ribosomal RNA gene and a large (16S) ribosomal RNA gene, 22 transfer RNAs and 13 protein-coding genes[3]. The mtDNA-encoded polypeptides are enzyme complex subunits of the oxidative phosphorylation (OXPHOS) system responsible for the synthesis of ATP[4]. In addition, mtDNA lacks introns, and is susceptible to reactive oxygen species (ROS)[5-7], and involved in carcinogenesis because of its high susceptibility to mutations and limited repair mechanisms in comparison to nuclear DNA[8-10]. Quantitative alteration of transcripts (mtRNA) of mtDNA may be a general characteristic of cancer cells[2]. Increased transcripts of mtDNA may lead to decreased apoptosis of tumor cells and subsequent carcinogenesis[11]. Microarray technique can monitor the expression of many genes in parallel, thereby speeding up the identification of differentially expressed genes and the construction of differential expression profiles. Microarray analysis has become an increasingly popular tool to study the functions of genes, especially those genes involved in tumor formation and growth[12-15].
One normal gastric mucosal tissue, 6 gastric cancer tissues and corresponding normal gastric epithelial mucosal tissues adjacent to cancerous lesions were obtained from the First Affiliated Hospital Hospital of China Medical University, which were all diagnosed pathologically by hematoxylin and eosin staining.
Primer designation Both the primer design software Oligo 6.22, Primer Premier 5.0 and Mitomap database (human mitochondrial genome database of Emory) were used to analyze the whole mitochondrial genome, and 15 pairs of primers were screened out to amplify the probes of mtRNA (Table 1). BLAST analysis was used to exclude the influence of nuclear pseudogenes.
| Functionaldomain | Primer | Sequence | Length (mer) | Tm ( °C) | GC% | Product size (bp) |
| 12SrRNA | F1097 | GCCCTAAACCTCAACAGT | 18 | 54.1 | 50 | 264 |
| R1360 | CATTTCTTGCCACCTCAT | 18 | 44.5 | 44.4 | ||
| 16SrRNA | F2618 | TAGGGACCTGTATGAATGG | 19 | 48.2 | 47.4 | 485s |
| R3102 | ATAGAAACCGACCTGGAT | 18 | 48 | 44.4 | ||
| ND1 | F3927 | GTCTCAGGCTCAACATC | 17 | 42.1 | 52.9 | 273 |
| R4199 | TAGGGTGAGTGGTAGGAA | 18 | 51.1 | 50 | ||
| ND2 | F5022 | CCCACATAGGATGAATAA | 18 | 40.2 | 38.9 | 466 |
| R5487 | GCGATGAGGATGGATAGAG | 19 | 46.2 | 52.6 | ||
| COI | F6043 | TCTAGGTAACGACCACATCTACAAC | 25 | 62.4 | 44 | 614 |
| R6656 | CGAAGCCTGGTAGGATAA | 18 | 51.9 | 50 | ||
| COII | F7841 | TAACAGACGAGGTCAACG | 18 | 50.5 | 50 | 351 |
| R8191 | TTGCTCCACAGATTTCAG | 18 | 43.1 | 44.4 | ||
| ATPase8 | F8366 | TGCCCCAACTAAATACTA | 18 | 49.4 | 38.9 | 191 |
| R8556 | CAATGAATGAAGCGAACA | 18 | 41.6 | 38.9 | ||
| ATPase6 | F9000 | CGCCTAACCGCTAACATTACTG | 22 | 64.2 | 50 | 148 |
| R9147 | AGGCGACAGCGATTTCTA | 18 | 53.8 | 50 | ||
| COIII | F9321 | CCATAACGCTCCTCATAC | 18 | 47.8 | 50 | 203 |
| R9523 | TAGGCTGGAGTGGTAAAA | 18 | 51.8 | 44.4 | ||
| ND3 | F10200 | GCGTCCCTTTCTCCATAA | 18 | 52.4 | 50 | 203 |
| R10402 | TTCGGTTCAGTCTAATCCTT | 20 | 51.8 | 40 | ||
| ND4 | F11581 | ATCTGCCTACGACAAACA | 18 | 48.3 | 44.4 | 444 |
| R12024 | GTGGTGGGTGAGTGAGCCC | 19 | 61.3 | 68.4 | ||
| ND4L | F10573 | AATAATACTATCGCTGTTCA | 19 | 45.4 | 30 | 189 |
| R10761 | CATTGGAGTAGGTTTAGG | 18 | 46.2 | 44.4 | ||
| ND5 | F13028 | CTGACTCCCCTCAGCCATAGA | 21 | 57.2 | 57.1 | 276 |
| R13303 | TGTGGTTGGTTGATGCCG | 18 | 53.3 | 55.6 | ||
| ND6 | F14322 | GTTTACCACAACCACCAC | 18 | 52.1 | 50 | 291 |
| R14612 | TCTAAGCCTTCTCCTATTT | 19 | 48.8 | 36.8 | ||
| Cyt-b | F15002 | GCGCCTCAATATTCTTTATCTGC | 23 | 58.7 | 43.5 | 305 |
| R15306 | GAAGGGCAAGATGAAGTGAAA | 21 | 53.4 | 42.9 |
RNA isolation Total RNA was extracted from normal gastric tissues, gastric cancerous and para-cancerous mucosal tissues. A total of 50-100 mg frozen tissue was pulverized in a mortar containing liquid nitrogen. The powder was dissolved in TRIzol reagent, and then chloroform was added to precipitate the protein. RNA was isolated by precipitation with isopropanol. RNA pellet was washed in 750 mL/L ethanol, air-dried and dissolved in water treated with diethylpyrocarbonate (DEPC). RNA was stored at -80 °C until use.
Probe preparation RNA extracted from normal gastric samples was amplified via reverse transcription PCR (RT-PCR) to produce the probes of mtRNA. Reverse transcription procedure referred to the manual of TaKaRa AMV RT-PCR kit. PCR reaction was carried out in a final volume of 50 μL in a Biometra personal PCR system, with an initial incubation at 94 °C for 4 min, followed by 30 cycles, each consisting of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 2 min. Finally a further extension was performed at 72 °C for 4 min. Meanwhile, two housekeeping genes β-actin (X16940) and glyceraldehyde phosphate dehydrogenase (GAPDH, M33197) and two hepatitis C virus (HCV) genes were amplified as the positive internal control and negative control, respectively. Polyacrylamide gel electrophoresis (PAGE) confirmed the integrity of the probes and control genes. Probes were purified according to the manual of QIAquick PCR purification kit and quantified with UV absorbance at 260 nm, finally re-suspended in 0.1× carbonate buffer (pH 9.0) or in an aqueous solution of 150 mmol/L sodium phosphate (pH 8.5) plus 0.1 g/L SDS[16].
RT-PCR of cDNA labeled with fluorescence One microgram of RNA extracted from gastric cancerous and para-cancerous tissues was reverse-transcribed into cDNA in 20 μL reaction volume with a hexamer primer (TaKaRa AMV RT-PCR Kit) and was labeled with fluorescein isothiacyanate dUTP (FITC-dUTP). To remove RNA complementary to cDNA, 1 μL of E.coli RNAase H was added and incubated at 37 °C for 20 min to digest the residue RNA. The resulting cDNA was resuspended in 20 μL deionized water, and stored at -20 °C.
Amino-slides Coverslides were soaked for over 24 h in the mixture of dichromic acid and stronger sulfuric acid, rinsed with tap water, and then plunged into 1 mol/L NaOH for 1 h. The slides were washed with an ultrasonic washing device for 3×3 min, and dipped in acetone for 3 min, in 50 mL/L arm element KH-550 (with acetone) for 6 min, in acetone for 5×3 min again, and then baked for 1 h at 104 °C.
Spotting and hybridization Fifteen pairs of mitochondrial DNA probes together with positive control housekeeping genes and negative control HCV gene were spotted onto amino-modified slides by a touching needle-dipping device (Micro Grid II device, England). To sufficiently analyze the results and preclude the interference of occasional errors, we spotted 9 spots per sample. The 9 spots were placed in a wet chamber with a humidity of 95% at 37 °C for 2 h, baked at 80 °C for 1 h, dipped in blocking solution (100 mL/L iodized skellysolve butane and 900 mL/L anhydrous alcohol) for 1.5 h.
Eight μL of RT-PCR products and 2μL of hybridization buffer containing 300 mL/L DMSO (dimathyl sulfoxide) and 700 mL/L 20×SC were mixed. The amino-modified slides with probes and cDNA mixture above were denatured respectively at 95 °C for 5 min, dipped quickly into ice-cold water for 3 min. The mixture was added onto the slides, and then the silicon-slide was placed on the top of the array to make them fully hybridized, the slides were placed in a well-sealed hybridization chamber, and incubated in a 55 °C oven for 12-14 h.
Slide washing The slides were washed in 0.5×SC/0.1 g/L SDS solution at 42 °C for 5 min and in 2×SC at 37 °C for 3 min with gentle agitation, stored in a lightproof slide box.
Detection Chips were scanned with a scanning array system at 10 μm resolution (GeneTACTM laser scanner, USA). The obtained images were analyzed using ImaGene3.0 software (BioDiscovery, Los Angeles, USA).
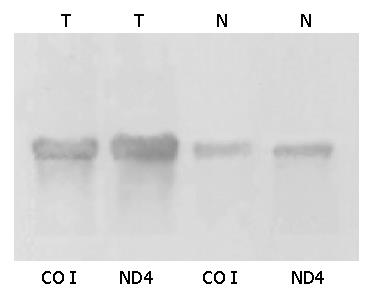
Northern blot In order to evaluate the reliability of the microarray method, the RNA extracted from gastric cancerous and non-cancerous tissues was subjected to Northern blot analysis. Probes of NADH dehydrogenase 4 (ND4), cytochrome C oxidase I (COI) were labeled with a random prime DNA labeling kit (Boehringer-Mannheim). Equal amounts of RNA determined by quantitation of optical densities at 260 nm and further normalized using the housekeeping genes, were loaded onto agarose gels containing 2.2 mol/L formaldehyde, and transferred to nylon membranes. The membranes were dried and prehybridized at 42 °C for 3 h, and then hybridized with labeled ND4 and COI at 42 °C for 18 h.
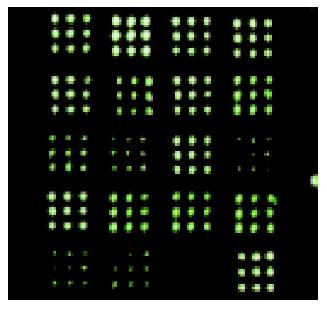
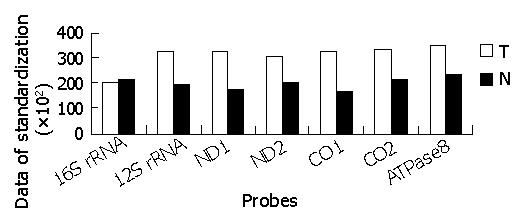
The hybridized spots were distinct, with a clear border and no black cavity, the background was consistent and clear for image analysis (Figure 1). In All arrays, the housekeeping genes showed positive signals, whereas HCV genes showed negative signals. The intensity of each spot represented the quantity of FITC-dUTP, hybridized to each spot. In order to enhance the confidence of the results, the overall intensities were normalized with a correction coefficient obtained using the ratios of housekeeping genes (Figure 2). After data were standardized, the results showed that the expression levels of some mitochondrial genes in gastric carcinoma were higher than those in the para-cancerous tissues. Since the samples were limited, further detailed analyses would be reported with a large size of samples.
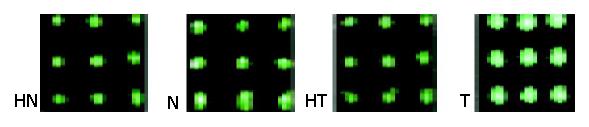
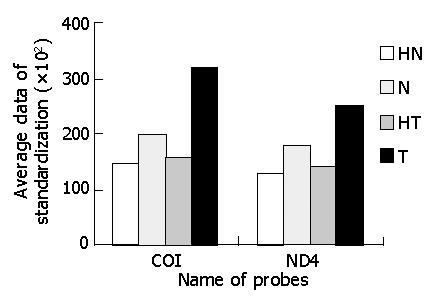
Northern blot showed that transcripts of some parts of mtDNA in cancerous tissues were higher than those in non-cancerous tissues, and increased differently in different parts of mtRNA. ND4 and COI were used to serve as two representatives of the whole probes of mtRNA (Figures 3,4 and 5). The Northern blot results displaying a high concordance with the microarray further indicated that the established microarray method was reliable.
Tumor cells, in general, contain fewer mitochondria than corresponding normal tissues[2,17]. The diminished content of mitochondria in tumor cells thus might reflect a reduced expression of mitochondrial or nuclear genes for mitochondrial proteins in response to neoplastic transformation[1,18]. However, paradoxically enhanced expression of mitochondrial genes in cancer has been reported[1,11,19]. In solid tumors, an elevated expression of mtDNA-encoded subunits of the mitochondrial electron respiratory chain might reflect mitochondrial adaptation to perturbations in cellular energy requirements[11]. Increased mtRNA levels might possibly suppress tumor cell apoptosis, and subsequently lead to the overgrowth of tumor cells[11]. The precise relationship between mitochondrial mass, level of mitochondrial mRNA, and mtDNA copy number has to be examined. Whether the levels of tumor cell mtDNA increase or decrease, specific alterations of mtDNA gene transcripts might be a tumor marker. Some differentially expressed gene profiles might accompany a specific or a type of carcinoma[20-22]. Hence, we can use the novel method to screen the sense parts of mtRNA as a tumor marker or even as a different tumor marker.
Some traditional methods can be applied to this work such as RT-PCR, Northern blot and real-time PCR, but they are time-consuming and rather expensive. A single microarray could provide information on the expression of tens of thousands of genes. The success of fully exploiting these powerful approaches depends on several criteria[23-27]: accurate selection, amplification and location of probe molecules, accurate reference sequence information, identification of unique probes, accurate distinction among multiple products of a single gene, accurate reconstruction of expressed sample nucleotide sequences, precision map scanning, and reproducible and accurate transformation of image files to numerical data. Expression analysis using glass slide microarrays is typically performed by the competitive hybridization of two targets (typically known as test and reference), each labeled with a specific fluorescent dye like FITC, Cy3 and Cy5. There are also a number of reasons why data must be normalized, including unequal quantities of starting RNA, differences in labeling or detection efficiencies between the fluorescent dyes used, and systematic biases in the measured expression levels[28-32]. Though we also could use two different fluoresceins to label gastric cancerous and normal tissues respectively, yet considering the above difficulties of microarray data normalization and map transformation, we only labeled one fluorescent FITC to gastric cancer and normal gastric tissues, and used the housekeeping gene to normalize the hybridization signals of mtRNA probes. In this study, in the course of constructing the microarray, We paid more attention to the good and consistent background of the microarrayt[33,34]. The processing procedure of slides is directly related to the data analysis. Moreover, since single point data can not draw exact data on account of accidental errors, spotting several spots is essential. Otherwise the results might be artificially positive or negative. In conclusion, mitochondrial microarray is a reliable and repeatable method to detect the loss or changes of mtDNA expression levels, but the precise mechanisms by which the two genomes interact and integrate with each other are poorly understood.
We thank the Biochip Center of China Medical University for providing the reagents and technical assistance.
| 1. | Luciaková K, Kuzela S. Increased steady-state levels of several mitochondrial and nuclear gene transcripts in rat hepatoma with a low content of mitochondria. Eur J Biochem. 1992;205:1187-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 2. | Penta JS, Johnson FM, Wachsman JT, Copeland WC. Mitochondrial DNA in human malignancy. Mutat Res. 2001;488:119-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 328] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 3. | Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410:103-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 995] [Cited by in RCA: 1098] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 4. | Bianchi NO, Bianchi MS, Richard SM. Mitochondrial genome instability in human cancers. Mutat Res. 2001;488:9-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 132] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 5. | Lee I, Bender E, Kadenbach B. Control of mitochondrial membrane potential and ROS formation by reversible phosphorylation of cytochrome c oxidase. Mol Cell Biochem. 2002;234-235:63-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 114] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 6. | Lee I, Bender E, Arnold S, Kadenbach B. New control of mitochondrial membrane potential and ROS formation--a hypothesis. Biol Chem. 2001;382:1629-1636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 80] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 7. | Atlante A, Calissano P, Bobba A, Azzariti A, Marra E, Passarella S. Cytochrome c is released from mitochondria in a reactive oxygen species (ROS)-dependent fashion and can operate as a ROS scavenger and as a respiratory substrate in cerebellar neurons undergoing excitotoxic death. J Biol Chem. 2000;275:37159-37166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 160] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 8. | Máximo V, Soares P, Seruca R, Rocha AS, Castro P, Sobrinho-Simões M. Microsatellite instability, mitochondrial DNA large deletions, and mitochondrial DNA mutations in gastric carcinoma. Genes Chromosomes Cancer. 2001;32:136-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 94] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 9. | Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20:291-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 625] [Cited by in RCA: 621] [Article Influence: 22.2] [Reference Citation Analysis (5)] |
| 10. | Han CB, Li F, Zhao YJ, Ma JM, Wu DY, Zhang YK, Xin Y. Variations of mitochondrial D-loop region plus downstream gene 1 2S rRNA-tRNA(phe) and gastric carcinomas. World J Gastroenterol. 2003;9:1925-1929. [PubMed] |
| 11. | Wang J, Silva JP, Gustafsson CM, Rustin P, Larsson NG. Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. Proc Natl Acad Sci U S A. 2001;98:4038-4043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 195] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 12. | Tefferi A, Bolander ME, Ansell SM, Wieben ED, Spelsberg TC. Primer on medical genomics. Part III: Microarray experiments and data analysis. Mayo Clin Proc. 2002;77:927-940. [PubMed] [DOI] [Full Text] |
| 13. | Thomas R, Fiegler H, Ostrander EA, Galibert F, Carter NP, Breen M. A canine cancer-gene microarray for CGH analysis of tumors. Cytogenet Genome Res. 2003;102:254-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Kerr MK. Design considerations for efficient and effective microarray studies. Biometrics. 2003;59:822-828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 128] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 15. | van den Boom J, Wolter M, Kuick R, Misek DE, Youkilis AS, Wechsler DS, Sommer C, Reifenberger G, Hanash SM. Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am J Pathol. 2003;163:1033-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 247] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 16. | Sterrenburg E, Turk R, Boer JM, van Ommen GB, den Dunnen JT. A common reference for cDNA microarray hybridizations. Nucleic Acids Res. 2002;30:e116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 51] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 17. | Han CB, Li F, Yang XF, Mao XY, Wu DY, Xin Y. Alterations of mtDNA copy number in gastric carcinoma. Shijie Huaren Xiaohua Zazhi. 2004;12:258-261. |
| 18. | Han CB, Zhao YJ, Li F, He Q, Ma JM, Xin Y. Quantitation and detection of deletion in tumor mitochondrial DNA by microarray technique. Zhonghua Zhong Liu Za Zhi. 2004;26:10-13. [PubMed] |
| 19. | Savre-Train I, Piatyszek MA, Shay JW. Transcription of deleted mitochondrial DNA in human colon adenocarcinoma cells. Hum Mol Genet. 1992;1:203-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 20. | Lu P, Nakorchevskiy A, Marcotte EM. Expression deconvolution: a reinterpretation of DNA microarray data reveals dynamic changes in cell populations. Proc Natl Acad Sci USA. 2003;100:10370-10375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 94] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 21. | Killion PJ, Sherlock G, Iyer VR. The Longhorn Array Database (LAD): an open-source, MIAME compliant implementation of the Stanford Microarray Database (SMD). BMC Bioinformatics. 2003;4:32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 60] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 22. | Alonso A, Mahmood R, Li S, Cheung F, Yoda K, Warburton PE. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum Mol Genet. 2003;12:2711-2721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 83] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 23. | Kendziorski CM, Zhang Y, Lan H, Attie AD. The efficiency of pooling mRNA in microarray experiments. Biostatistics. 2003;4:465-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 160] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 24. | El-Bayoumy K, Narayanan BA, Desai DH, Narayanan NK, Pittman B, Amin SG, Schwartz J, Nixon DW. Elucidation of molecular targets of mammary cancer chemoprevention in the rat by organoselenium compounds using cDNA microarray. Carcinogenesis. 2003;24:1505-1514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 28] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 25. | Broekhuijsen M, Larsson P, Johansson A, Byström M, Eriksson U, Larsson E, Prior RG, Sjöstedt A, Titball RW, Forsman M. Genome-wide DNA microarray analysis of Francisella tularensis strains demonstrates extensive genetic conservation within the species but identifies regions that are unique to the highly virulent F. tularensis subsp. tularensis. J Clin Microbiol. 2003;41:2924-2931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 148] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 26. | Ramos-Nino ME, Scapoli L, Martinelli M, Land S, Mossman BT. Microarray analysis and RNA silencing link fra-1 to cd44 and c-met expression in mesothelioma. Cancer Res. 2003;63:3539-3545. [PubMed] |
| 27. | Song JW, Lin JS, Kong XJ, Liang KH. Clinical study of oligonucleotide microarray on monitoring the lamivudine-resistance mutations in hepatitis B virus. Zhonghua GanZangBing ZaZhi. 2003;11:361-363. [PubMed] |
| 28. | Hasegawa S, Furukawa Y, Li M, Satoh S, Kato T, Watanabe T, Katagiri T, Tsunoda T, Yamaoka Y, Nakamura Y. Genome-wide analysis of gene expression in intestinal-type gastric cancers using a complementary DNA microarray representing 23,040 genes. Cancer Res. 2002;62:7012-7017. [PubMed] |
| 29. | Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19:1236-1242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 274] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 30. | Yoo GH, Piechocki MP, Ensley JF, Nguyen T, Oliver J, Meng H, Kewson D, Shibuya TY, Lonardo F, Tainsky MA. Docetaxel induced gene expression patterns in head and neck squamous cell carcinoma using cDNA microarray and PowerBlot. Clin Cancer Res. 2002;8:3910-3921. [PubMed] |
| 31. | Hariharan R. The analysis of microarray data. Pharmacogenomics. 2003;4:477-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 32. | Mantripragada KK, Buckley PG, Jarbo C, Menzel U, Dumanski JP. Development of NF2 gene specific, strictly sequence defined diagnostic microarray for deletion detection. J Mol Med (Berl). 2003;81:443-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 33. | Abe S, Katagiri T, Saito-Hisaminato A, Usami S, Inoue Y, Tsunoda T, Nakamura Y. Identification of CRYM as a candidate responsible for nonsyndromic deafness, through cDNA microarray analysis of human cochlear and vestibular tissues. Am J Hum Genet. 2003;72:73-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 98] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 34. | Campanaro S, Romualdi C, Fanin M, Celegato B, Pacchioni B, Trevisan S, Laveder P, De Pittà C, Pegoraro E, Hayashi YK. Gene expression profiling in dysferlinopathies using a dedicated muscle microarray. Hum Mol Genet. 2002;11:3283-3298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 63] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
Edited by Wang XL and Kumar M