Published online Apr 15, 2004. doi: 10.3748/wjg.v10.i8.1183
Revised: November 14, 2003
Accepted: December 16, 2003
Published online: April 15, 2004
AIM: To construct a prokaryotic expression system of a Helicobacter pylori (H pylori) cagA gene fragment and establish enzyme-linked immunosorbent assays (ELISA) for detecting CagA and its antibody, so as to understand the manner in which the infection of CagA-expressing H pylori (CagA+H pylori) isolates cause diseases.
METHODS: H pylori strains in gastric biopsy specimens from 156 patients with positive results in rapid urease test were isolated. PCR was used to detect the frequency of cagA gene in the 109 H pylori isolates and to amplify a 2 148-bp fragment (cagA1) of cagA gene from a clinical strain Y06. A prokaryotic expression system of cagA1 gene was constructed, and the expression of the target recombinant protein (rCagA1) was examined by SDS-PAGE. Western blotting and immunodiffusion assay were employed to determine the immunoreactivity and antigenicity of rCagA1, respectively. Two ELISAs were established to detect CagA expression in 109 H pylori isolates and the presence of CagA antibody in the corresponding patients’ sera, and the correlations between infection with CagA+H pylori and gastritis as well as peptic ulcer were analyzed.
RESULTS: Of all the clinical specimens obtained, 80.8% (126/156) were found to have H pylori isolates and 97.2% of the isolates (106/109) were positive for cagA gene. In comparison with the reported data, the cloned cagA1 fragment possessed 94.83% and 93.30% homologies with the nucleotide and putative amino acid sequences, respectively. The output of rCagA1 produced by the constructed recombinant prokaryotic expression system was approximately 30% of the total bacterial protein. rCagA1 was able to bind to the commercial antibody against the whole-cells of H pylori and to induce the immunized rabbits to produce antibody with an immunodiffusion titer of 1:4. A proportion as high as 92.6% of the H pylori isolates (101/109) expressed CagA and 88.1% of the patients’ serum samples (96/109) were CagA antibody-positive. The percentage of CagA+H pylori strains (97.9%) isolated from the biopsy specimens of peptic ulcer appeared to be higher than that from gastritis (88.5%), but the difference was not statistically significant (χ2 = 3.48, P > 0.05).
CONCLUSION: rCagA1 produced by the prokaryotic expression system constructed in this study possesses good immunoreactivity and antigenicity, and the established ELISAs can be used to detect CagA of H pylori and its antibody. H pylori isolates show high frequencies of cagA gene and CagA expression, but the infections by CagA+H pylori strains are not the most decisive factors to cause gastric diseases.
-
Citation: Yan J, Wang Y, Shao SH, Mao YF, Li HW, Luo YH. Construction of prokaryotic expression system of 2 148-bp fragment from
cagA gene and detection ofcagA gene, CagA protein inHelicobacter pylori isolates and its antibody in sera of patients. World J Gastroenterol 2004; 10(8): 1183-1190 - URL: https://www.wjgnet.com/1007-9327/full/v10/i8/1183.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i8.1183
In China, gastritis and peptic ulcer are the most prevalent gastric diseases, and gastric cancer remains one of the most devastating malignant tumors with the highest morbidity[1-20]. Helicobacter pylori (H pylori) has been recognized as a human-specific gastric pathogen that colonizes in the stomach of at least half of the world’s populations[21,22]. Most infected individuals are asymptomatic, whereas in some cases, the infection causes acute, chronic gastritis or peptic ulceration, and plays an important role in the development of peptic ulcer and gastric adenocarcinoma, mucosa-associated lymphoid tissue (MALT) lymphoma and primary gastric non-Hodgkin’s lymphoma[23-28].
So far, no evidence for the toxicity of the protein (CagA) expressed by cytotoxin-associated gene A (cagA) of H pylori has been presented[29,30]. However, previous studies demonstrated that CagA was closely associated with the pathogenicity of H pylori and severity of H pylori-related diseases[29-32]. Many epidemiological data indicated that the positive rate of cagA gene was significantly higher in the H pylori strains isolated from patients with peptic ulcer than in those with gastritis[33]. Patients infected with cagA+H pylori had a higher risk of developing gastric cancers than those infected with cagA-strains[34,35]. Approximately 60% to 70% of H pylori strains isolated from European and North American populations carried cagA gene[36-38], whereas over 90% of the isolates from Asia-Pacific populations were cagA gene-positive[39-42]. Strong antigenicity of CagA usually induces antibody in patients with cagA+H pylori infection and this antibody has been considered as a possible specific clinical indicator of H pylori infection[43-45]. However, the data are scarce concerning the correlations between the presence of cagA, CagA expression and antibody production, CagA+H pylori infection and types of the resultant gastric diseases.
In the present study, a recombinant expression plasmid containing a relatively conserved H pylori cagA gene fragment 2 148 bp in length (cagA1) was constructed. H pylori strains in gastric biopsy specimens from patients with gastritis or peptic ulcer were isolated. The frequencies of cagA gene and expressions of H pylori isolates and CagA antibody in patients’ sera were investigated. Furthermore, the correlations among CagA+H pylori infection and types of the resulted gastric diseases were also analyzed for the purpose of understanding the pathogenic effect of CagA and the potential of CagA antibody detection in clinical diagnosis of H pylori infection.
A typical H pylori strain named Y06 isolated clinically was used to amplify cagA1 fragment. Primers for PCR amplification were synthesized by BioAsia (Shanghai, China). Taq-plus high fidelity PCR kit and restriction endonucleases were purchased from TaKaRa (Dalian, China). T-A cloning kit and sequencing service were provided by BBST (Shanghai, China). Plasmid pET32a as the expression vector and E. coli BL21DE3 as the host cell were purchased from Novagen (Novagen, Madison, UNITED STATES). Rabbit antiserum against the whole cell of H pylori, HRP-labeling sheep antisera against rabbit IgG and human IgG were purchased from DAKO (Glostrup, Denmark) and Jackson ImmunoResearch (West Grove, UNITED STATES), respectively. Agents used for isolation and identification of H pylori were purchased from BioMérieux (Marcy I’Etoile, France).
Gastric biopsy specimens with positive urease for H pylori isolation and serum samples for CagA antibody detection were collected from 156 patients in 4 hospitals in Hangzhou during Nov. 2001 to Feb. 2003. None of the patients had received nonsteroidal anti-inflammatory drugs, antacids or antibiotics within two weeks prior to the study. From the 156 patients, 126 biopsy specimens of H pylori isolates were obtained and 109 of the isolates survived after -70 °C storage. In the 109 patients (including 76 males and 33 females, with a mean age of 40.2 years), 61 patients suffered from chronic gastritis (41 chronic superficial gastritis, 10 chronic active gastritis, 10 chronic atrophy gastritis), and 48 had gastroduodenal ulcer (12 gastric ulcer, 30 duodenal ulcer, 6 gastric and duodenal ulcer).
Isolation and identification of H pylori Each of the biopsy specimen was homogenized with a tissue grinder and then inoculated on Columbia agar plates supplemented with 80 mL/L sheep blood, 5 g/L cyclodextrin, 5 mg/L trimethoprim, 10 mg/L vancomycin, 2.5 mg/L amphotericin B and 2 500 U/L cefsoludin. The plates were incubated at 37 °C under microaerobic conditions (50 mL/L O2, 100mL/L CO2 and 850 mL/L N2) for 3 to 5 d. A bacterial isolate was identified as H pylori according to typical Gram staining morphology, positive results of biochemical tests for urease and oxidase, and slide agglutination with the commercial rabbit antibody against whole cell of the bacterium.
Preparation of DNA template Genomic DNA from each of the H pylori strains was extracted by conventional phenol-chloroform method and DNase-free RNase treatment. Concentration and purity of the DNA preparations were determined by ultraviolet spectrophotometry[46].
Polymerase Chain reaction The primers were designed to amplify cagA1 fragment from H pylori strain Y06 cagA gene based on the published data in GenBank. The sequence of sense primer with an endonuclease site of EcoR V was 5’-GCCGAT ATCATGATCAATAATCTTCAAGTAGC-3’, and that of the antisense primer with an endonuclease site of XhoI was 5’-CGGCTCGAGTTATGAAAT CCATTCTGGATTG-3’. The total volume per PCR was 100 µL, containing 2.5 mol/L each of the dNTP, 250 nmol/L each of the two primers, 15 mol/L MgCl2, 3.0 U Taq-plus polymerase, 100 ng DNA template and 1 × PCR buffer (pH 8.3). The parameters for PCR were at 94 °C for 5 min, × 1; at 94 °C for 30 s, at 50 °C for 30 s, at 72 °C for 120 s, × 10; at 94 °C for 30 s, at 50 °C for 30 s, at 72 °C for 130 s (additional 10 s for each of the following cycles), × 20; at 72 °C for 10 min, × 1.
To increase the positive detection rate of cagA gene, two sets of primers derived from different regions of cagA gene were applied in PCR. The sequences of F1/B1 primers were 5’-GATAACAGGCAAGCTTTTGAGG-3’(sense), 5’-CTGCAAAAGATTGTTTGGCAGA-3’(antisense)[31]. The sequences of D008/R008 primers were 5’-ATAATGCTAAAT TAGACAACTTGAGCG-3’(sense), 5’-TTAGAATAATCAA CAAACATCACGCCA-3’ (antisense)[36]. Except for the primers and DNA templates, all the other reagents and reaction volumes used in PCR for cagA detection were the same as in cagA1 amplification. The parameters for the two PCRs were at 94 °C for 5 min, × 1; at 94 °C for 30 s, at 55 °C for 1 min, at 72 °C for 90 s, × 35; at 72 °C for 7 min, × 1.
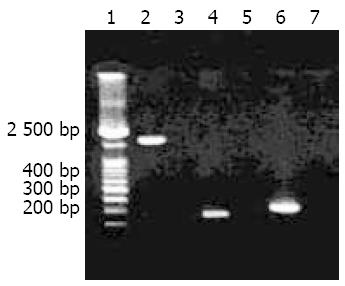
The results of PCR were observed under UV light after electrophoresis on 1.5% agarose gel pre-stained with ethidium bromide. The expected sizes of cagA1 amplification fragment and the two target amplification fragments for cagA gene detection were 2 172 bp (including ATG, TAA, and a 18-bp sequence containing endonuclease sites and protective nucleotide residuals), 349 bp and 298 bp, respectively.
Cloning and sequencing The cagA1 amplification fragment was cloned into plasmid vector pUCm-T (pUCm-T-cagA1) by using the T-A cloning kit according to the manufacturer’s instructions. The recombinant plasmid was amplified in E. coli DH5α and then extracted by Sambrook’s method[46]. A professional company (BBST) was responsible for nucleotide sequence analysis of the inserted fragment. Two plasmids pUCm-T-cagA1 and pET32a were extracted from two different strains of E. coli DH5α after amplification in LB medium and then digested with EcoRV and XhoI, respectively[46]. The fragments cagA1 and pET32a were recovered and ligased. The recombinant expression vector pET32a- cagA1 was transformed into E. coli BL21DE3, and the expression system designated as pET32a-cagA1-E. coli BL21DE3. The cagA1 fragment inserted in pET32a was sequenced again.
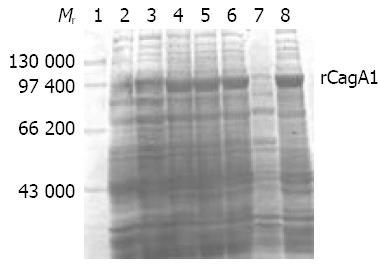
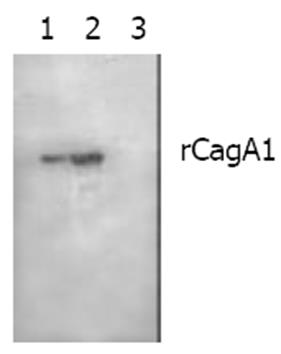
Expression and identification of target recombinant proteinpET32a-cagA1-E. coli BL21DE3 was rotatively cultured in LB medium at 37 °C under induction with isopropylthio-β-D-galactoside (IPTG) at different concentrations of 1.0, 0.5 and 0.1 mmol/L, respectively. The supernatant and precipitate of the culture after incubation were separated by centrifugation and then the bacterial pallets were ultrasonically fragmented (300 V, 5 s × 3). SDS-PAGE was used to measure the molecular mass and output of the target recombinant protein (rCagA1). Ni-NTA affinity chromatography was applied to collect rCagA1. The commercial rabbit antiserum against whole-cell H pylori and HRP-labeling sheep antiserum against rabbit IgG were used as the first and second antibodies to determine the immunoreactivity of rCagA1 by Western blotting, respectively. Rabbits were immunized with rCagA1 to prepare antisera. Immunodiffusion assay was performed to determine the antigenicity of rCagA1.
Enzyme-linked immunosorbent assay (ELISA) By using rCagA1 as the coating antigen at the concentration of 20 µg/mL, with the serum sample (1:400 dilution) from a patient as the first antibody and HRP-labeling sheep antibody against human IgG (1:4 000 dilution) as the second antibody, CagA antibody in the sera of the 126 H pylori-infected patients was detected. The result of ELISA for a patient’s serum sample was considered positive if the value of optical density at 490 nm (OD490) exceeded the mean plus 3 standard deviations of 6 different negative serum samples[47]. CagA expression in H pylori isolates was examined using ultrasonic supernatant of each of the H pylori isolates (50 µg/mL) as the coating antigen, self-prepared rabbit anti-rCagA1 serum (1:800 dilution) as the first antibody and HRP-labeling sheep antibody against rabbit IgG (1:3 000 dilution) as the second antibody. The result of ELISA for a H pylori ultrasonic supernatant sample was considered positive if its OD490 value was over the mean plus 3 standard deviations of 6 separated E. coli DH5α ultrasonic supernatant samples at the same protein concentration[47].
Analysis of correlation among cagA gene, CagA and its antibody and H pylori-related diseases According to the clinical data and the obtained results, the correlations among infection with H pylori carrying cagA gene and expressing CagA isolated from the patients’ gastric biopsy specimens, and the type and severity of gastric diseases in the same patient were analyzed.
The nucleotide and putative amino acid sequences of the cloned cagA1 fragment were compared for homologies with the published corresponding sequences (GenBank accession No. AB015416). χ2 test was applied to analyze the clinical data, PCR results for cagA detection and ELISA results for CagA detection.
In the 156 gastric biopsy specimens with positive urease, H pylori was detectable in 126 specimens, with a positivity rate of 80.8%.
Using the primer pairs F1/B1 and D008/R008 respectively, 82.6% and 78.9% of the tested H pylori isolates (90/109) were positive for cagA gene, and the total cagA gene positivity rate was 97.2%(106/109). The target amplification products of cagA1 from H pylori strain Y06 and two fragments for cagA gene detection from the isolates are shown in Figure 1.
The nucleotide sequences of cagA1 fragment in pUCm-T-cagA1 and pET32a-cagA1 were completely the same. The nucleotide and putative amino acid sequences of the cloned cagA1 fragment showed 94.83% (Figure 2) and 93.30% (Figure 3) homologies with the published sequences from H pylori strain NCTC11637 (GenBank accession No.: AB015416), respectively.
IPTG at concentrations of 1.0, 0.5 and 0.1 mmol/L could efficiently induce the expression of rCagA1, which was detected mainly in the ultrasonic precipitate with an output of approximately 30% of the total bacterial proteins (Figure 4).
The commercial rabbit antibody against whole-cell H pylori could bind to rCagA1 as confirmed by Western blotting (Figure 5). Immunodiffusion assay demonstrated a titer of 1:4 between rCagA1 and rabbit anti-rCagA1 serum.
The mean A490 values (mean ± SD) of the 6 negative serum samples was 0.37 ± 0.03 in the detection of specific antibodies in sera of patients, and the positive reference value of 0.46 was consequently derived. According to the reference value, 88.1% (96/109) of the tested patients’ serum samples were positive for the rCagA1 antibodies with an A490 value ranging from 0. 56 to 1.05 (Table 1). From the mean A490 values of the 5 negative bacterial controls (0.27 ± 0.09) in the detection of CagA1 in H pylori isolates, the positive reference value of 0.54 was derived. According to the reference value, the epitope of rCagA1 with an A490 value ranging from 0.55% to 0.9792.6%(101/109) was detected in the tested H pylori isolates (Table 1).
| Tested indicator | Tested | Positive | Negative | Positivity |
| cases | cases | cases | rate (%) | |
| CagA gene | 109 | 106 | 3 | 97.2 |
| CagA protein | 109 | 101 | 8 | 92.6 |
| Anti-CagA | 109 | 96 | 13 | 88.1 |
A high rate of 97.9% of H pylori isolated from the peptic ulcer specimens (47/48) and 88.5% from the gastritis specimens (54/61) was positive for CagA expression, showing no statistically significant difference between the two positive rates (χ2 = 3.48, P > 0.05). Of the 8 H pylori strains in which CagA expression failed to be detected, 1, 1 and 6 strains were isolated from the biopsy specimens of gastric ulcer, chronic active gastritis and chronic superficial gastritis, respectively, and this distribution did not show any statistically significant difference, either (χ2 = 1.23, P > 0.05).
CagA expressed by H pylori was demonstrated to induce cellular skeleton rearrangement and interleukin (IL)-8 secretion in gastric epithelial cells[31,32]. IL-8, recognized as an inflammatory cytokine, could cause inflammation by inducing gathering of neutrophilic cells[48,49]. Infection with cagA+H pylori may elevate the risks of atrophic gastritis, intestinal metaplasia and gastric adenocarcinoma[34,50,51], and CagA is therefore considered as the most important pathogenic factor of H pylori.
CagA gene had a single copy located at the terminal end of region I in cag pathogenic island (CPI)[31,52], and was prone to mutation, especially in the 3’-end by insertion of different numbers of repeated sequences, resulting in the great variation in its length ranging from 3 444 to 5 925 bp in different isolates[36,37]. According to the analysis of 37 cagA gene sequences from GenBank, a fragment with approximate 65 bp starting from the 5’-end of cagA gene of different H pylori isolates also exhibited frequent mutations such as replacement, insertion and deletion, etc. Therefore, a relatively conserved fragment of 2 148 bp from the 67th to 2 214th bp at 5’-end of cagA gene was selected for cloning, which was provisionally named as cagA1. In this study, homologies of the nucleotide and amino acid sequences of the cloned cagA1 fragment reached 94.83% and 93.30% respectively in comparison with the reported sequences in GenBank (accession No.AB015416). High output of rCagA1, approximately 30% of the total bacterial protein, expressed by the constructed recombinant prokaryotic expression system pET32a-cagA1-E. coli BL21DE3 was confirmed by SDS-PAGE. rCagA1 could be recognized by a commercial antibody against whole-cell H pylori and was able to induce rabbit to produce high-titer antibodies, indicating that rCagA1 with good immunoreactivity and antigenicity can be used in ELISA as a qualified antigen for detecting CagA antibody and for preparing animal antiserum to detect CagA.
Yang et al reported that all the cagA+H pylori isolates were capable of expressing CagA[41]. However, we found that 5 strains of cagA+H pylori (7.3%) failed to exhibit the expression. Considering the highly likely mutation in cagA genes from different H pylori isolates, this non-expression of CagA was probably due to sequence mutation or abnormal transcription and translation[36,37]. In the present study, 88.1% of the serum samples from the H pylori-infected patients (96/109) were positive for CagA antibody, and the positive rate was only slightly lower than those of cagA gene (97.2%) and CagA (92.6%) of the isolates, suggesting that CagA possesses strong antigenicity and usually induces detectable specific antibody in cagA+ H pylori-infected patients. However, we found in this study that 11.9% of the serum samples were negative for CagA antibody (13/109), including 3 cases (2.8%) of cagA-H pylori infection, 5(4.6%) of cagA+H pylori infection and 5(4.6%) of CagA+H pylori infection. In addition, previously published data and our results suggest that over 90% of the H pylori isolates from Asia-Pacific areas were cagA gene-positive[39-42], whereas 60% to 70% of the H pylori isolates from European and North American areas were positive[36-38], indicating that the presence of CagA antibody could be used as a reference indicator with only a small risk of error for detecting H pylori infection in individuals from Asia-Pacific areas, but not for those from European or North American areas. It should be noted that the positive rate of cagA gene in the H pylori isolates in this study was as low as 78.9% to 82.6%, as detected using a single pair of primers F1/B1 or D008/R008, indicating that using multiple pairs of primers in PCR may increase the positive rate for cagA gene detection.
Covacci et al and Figueiredo et al reported that cagA+ H pylori infection could usually cause serious gastric diseases[37,38]. For example, 90% of H pylori isolates from peptic ulcer patients were cagA gene-positive, while only 50% to 60% of the isolates from superficial gastritis were positive. However, the reports from Asia-Pacific areas did not show a definite correlation between cagA+ H pylori infection and severity of the diseases[39-42]. Although cagA+ H pylori was isolated from chronic gastritis patients at a higher rate (97.9%) than from peptic ulcer patients (88.5%), the difference was not statistically significant (χ2 = 3.48, P > 0.05), probably due to the high rate of cagA gene-carrying H pylori (97.2%) and a relative small population tested in this study.
We are grateful to the four hospitals in Hangzhou that provided gastric biopsy specimens for this study, and helped us to complete this research.
| 1. | Zhang Z, Yuan Y, Gao H, Dong M, Wang L, Gong YH. Apoptosis, proliferation and p53 gene expression of H pylori associated gastric epithelial lesions. World J Gastroenterol. 2001;7:779-782. [PubMed] |
| 2. | Lu XL, Qian KD, Tang XQ, Zhu YL, Du Q. Detection of H.pylori DNA in gastric epithelial cells by in situ hybridization. World J Gastroenterol. 2002;8:305-307. [PubMed] |
| 3. | Yao YL, Xu B, Song YG, Zhang WD. Overexpression of cyclin E in Mongolian gerbil with Helicobacter pylori-induced gastric precancerosis. World J Gastroenterol. 2002;8:60-63. [PubMed] |
| 4. | Guo DL, Dong M, Wang L, Sun LP, Yuan Y. Expression of gastric cancer-associated MG7 antigen in gastric cancer, precancerous lesions and H pylori -associated gastric diseases. World J Gastroenterol. 2002;8:1009-1013. [PubMed] |
| 5. | Harry XH. Association between Helicobacter pylori and gastric cancer: current knowledge and future research. World J Gastroenterol. 1998;4:93-96. [PubMed] |
| 6. | Tu SP, Zhong J, Tan JH, Jiang XH, Qiao MM, Wu YX, Jiang SH. Induction of apoptosis by arsenic trioxide and hydroxy camptothecin in gastriccancer cells in vitro. World J Gastroenterol. 2000;6:532-539. [PubMed] |
| 7. | Cai L, Yu SZ, Zhang ZF. Helicobacter pylori infection and risk of gastric cancer in Changle County,Fujian Province,China. World J Gastroenterol. 2000;6:374-376. [PubMed] |
| 8. | Xu AG, Li SG, Liu JH, Gan AH. Function of apoptosis and expression of the proteins Bcl-2, p53 and C-myc in the development of gastric cancer. World J Gastroenterol. 2001;7:403-406. [PubMed] |
| 9. | Wang X, Lan M, Shi YQ, Lu J, Zhong YX, Wu HP, Zai HH, Ding J, Wu KC, Pan BR. Differential display of vincristine-resistance-related genes in gastric cancer SGC7901 cell. World J Gastroenterol. 2002;8:54-59. [PubMed] |
| 10. | Liu JR, Li BX, Chen BQ, Han XH, Xue YB, Yang YM, Zheng YM, Liu RH. Effect of cis-9, trans-11-conjugated linoleic acid on cell cycle of gastric adenocarcinoma cell line (SGC-7901). World J Gastroenterol. 2002;8:224-229. [PubMed] |
| 11. | Niu WX, Qin XY, Liu H, Wang CP. Clinicopathological analysis of patients with gastric cancer in 1200 cases. World J Gastroenterol. 2001;7:281-284. [PubMed] |
| 12. | Fang DC, Yang SM, Zhou XD, Wang DX, Luo YH. Telomere erosion is independent of microsatellite instability but related to loss of heterozygosity in gastric cancer. World J Gastroenterol. 2001;7:522-526. [PubMed] |
| 13. | Morgner A, Miehlke S, Stolte M, Neubauer A, Alpen B, Thiede C, Klann H, Hierlmeier FX, Ell C, Ehninger G. Development of early gastric cancer 4 and 5 years after complete remission of Helicobacter pylori associated gastric low grade marginal zone B cell lymphoma of MALT type. World J Gastroenterol. 2001;7:248-253. [PubMed] |
| 14. | Deng DJ. progress of gastric cancer etiology: N-nitrosamides 1999s. World J Gastroenterol. 2000;6:613-618. [PubMed] |
| 15. | Liu ZM, Shou NH, Jiang XH. Expression of lung resistance protein in patients with gastric carcinoma and its clinical significance. World J Gastroenterol. 2000;6:433-434. [PubMed] |
| 16. | Cai L, Yu SZ, Ye WM, Yi YN. Fish sauce and gastric cancer: an ecological study in Fujian Province,China. World J Gastroenterol. 2000;6:671-675. [PubMed] |
| 17. | Xue FB, Xu YY, Wan Y, Pan BR, Ren J, Fan DM. Association of H pylori infection with gastric carcinoma: a Meta analysis. World J Gastroenterol. 2001;7:801-804. [PubMed] |
| 18. | Wang RT, Wang T, Chen K, Wang JY, Zhang JP, Lin SR, Zhu YM, Zhang WM, Cao YX, Zhu CW. Helicobacter pylori infection and gastric cancer: evidence from a retrospective cohort study and nested case-control study in China. World J Gastroenterol. 2002;8:1103-1107. [PubMed] |
| 19. | Liu DH, Zhang XY, Fan DM, Huang YX, Zhang JS, Huang WQ, Zhang YQ, Huang QS, Ma WY, Chai YB. Expression of vascular endothelial growth factor and its role in oncogenesis of human gastric carcinoma. World J Gastroenterol. 2001;7:500-505. [PubMed] |
| 20. | Cao WX, Ou JM, Fei XF, Zhu ZG, Yin HR, Yan M, Lin YZ. Methionine-dependence and combination chemotherapy on human gastric cancer cells in vitro. World J Gastroenterol. 2002;8:230-232. [PubMed] |
| 21. | Michetti P, Kreiss C, Kotloff K, Porta N, Blano JL, Bachmann D, Herranz M, Saldinger PF, Corthesy-Theulaz I, Losonsky G. Oral immunization with urease and Escherichia coli heat-labile enterotoxin is safe and immunogenic in Helicobacter pylori-infected adults. Gastroenterology. 1999;116:804-812. [RCA] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 233] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 22. | Rollan A, Giancaspero R, Fuster F, Acevedo C, Figueroa C, Hola K, Schulz M, Duarte I. The long-term reinfection rate and the course of duodenal ulcer disease after eradication of Helicobacter pylori in a developing country. Am J Gastroenterol. 2000;95:50-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 23. | Suganuma M, Kurusu M, Okabe S, Sueoka N, Yoshida M, Wakatsuki Y, Fujiki H. Helicobacter pylori membrane protein 1: a new carcinogenic factor of Helicobacter pylori. Cancer Res. 2001;61:6356-6359. [PubMed] |
| 24. | Nakamura S, Matsumoto T, Suekane H, Takeshita M, Hizawa K, Kawasaki M, Yao T, Tsuneyoshi M, Iida M, Fujishima M. Predictive value of endoscopic ultrasonography for regression of gastric low grade and high grade MALT lymphomas after eradication of Helicobacter pylori. Gut. 2001;48:454-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 187] [Article Influence: 7.5] [Reference Citation Analysis (9)] |
| 25. | Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3126] [Cited by in RCA: 3246] [Article Influence: 129.8] [Reference Citation Analysis (1)] |
| 26. | Morgner A, Miehlke S, Fischbach W, Schmitt W, Müller-Hermelink H, Greiner A, Thiede C, Schetelig J, Neubauer A, Stolte M. Complete remission of primary high-grade B-cell gastric lymphoma after cure of Helicobacter pylori infection. J Clin Oncol. 2001;19:2041-2048. [PubMed] |
| 27. | Kate V, Ananthakrishnan N, Badrinath S. Effect of Helicobacter pylori eradication on the ulcer recurrence rate after simple closure of perforated duodenal ulcer: retrospective and prospective randomized controlled studies. Br J Surg. 2001;88:1054-1058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Gao HJ, Yu LZ, Bai JF, Peng YS, Sun G, Zhao HL, Miu K, L XZ, Zhang XY, Zhao ZQ. Multiple genetic alterations and behavior of cellular biology in gastric cancer and other gastric mucosal lesions: H.pylori infection, histological types and staging. World J Gastroenterol. 2000;6:848-854. [PubMed] |
| 29. | Atherton JC, Cao P, Peek RM, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771-17777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1078] [Cited by in RCA: 1113] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 30. | Stein M, Rappuoli R, Covacci A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci USA. 2000;97:1263-1268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 444] [Cited by in RCA: 451] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 31. | Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648-14653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1375] [Cited by in RCA: 1403] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 32. | Nogueira C, Figueiredo C, Carneiro F, Gomes AT, Barreira R, Figueira P, Salgado C, Belo L, Peixoto A, Bravo JC. Helicobacter pylori genotypes may determine gastric histopathology. Am J Pathol. 2001;158:647-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 130] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 33. | Peek RM, Moss SF, Tham KT, Pérez-Pérez GI, Wang S, Miller GG, Atherton JC, Holt PR, Blaser MJ. Helicobacter pylori cagA+ strains and dissociation of gastric epithelial cell proliferation from apoptosis. J Natl Cancer Inst. 1997;89:863-868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 225] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 34. | Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, Stemmermann GN, Nomura A. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111-2115. [PubMed] |
| 35. | Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut. 1997;40:297-301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 654] [Cited by in RCA: 673] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 36. | Tummuru MK, Cover TL, Blaser MJ. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect Immun. 1993;61:1799-1809. [PubMed] |
| 37. | Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791-5795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 883] [Cited by in RCA: 938] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 38. | Figueiredo C, Van Doorn LJ, Nogueira C, Soares JM, Pinho C, Figueira P, Quint WG, Carneiro F. Helicobacter pylori genotypes are associated with clinical outcome in Portuguese patients and show a high prevalence of infections with multiple strains. Scand J Gastroenterol. 2001;36:128-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 93] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 39. | Ito Y, Azuma T, Ito S, Miyaji H, Hirai M, Yamazaki Y, Sato F, Kato T, Kohli Y, Kuriyama M. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J Clin Microbiol. 1997;35:1710-1714. [PubMed] |
| 40. | Park SM, Park J, Kim JG, Yoo BC. Relevance of vacA genotypes of Helicobacter pylori to cagA status and its clinical outcome. Korean J Intern Med. 2001;16:8-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 41. | Yang H, Wu SV, Pichuantes S, Song M, Wang J, Zhou D, Xu Z, Quan S, Polito A, Walsh JH. High prevalence of cagA-positive strains in Helicobacter pylori-infected, healthy, young Chinese adults. J Gastroenterol Hepatol. 1999;14:476-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 42. | Park SM, Park J, Kim JG, Cho HD, Cho JH, Lee DH, Cha YJ. Infection with Helicobacter pylori expressing the cagA gene is not associated with an increased risk of developing peptic ulcer diseases in Korean patients. Scand J Gastroenterol. 1998;33:923-927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 23] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 43. | Chmiela M, Wiśniewska M, Bak-Romaniszyn L, Rechciński T, Płaneta-Małecka I, Bielański W, Konturek SJ, Płonka M, Klink M, Rudnicka W. Serological differentiation of Helicobacter pylori CagA(+) and CagA(-) infections. Arch Immunol Ther Exp (Warsz). 2003;51:131-136. [PubMed] |
| 44. | Park CY, Kwak M, Gutierrez O, Graham DY, Yamaoka Y. Comparison of genotyping Helicobacter pylori directly from biopsy specimens and genotyping from bacterial cultures. J Clin Microbiol. 2003;41:3336-3338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 45. | Tomasini ML, Zanussi S, Sozzi M, Tedeschi R, Basaglia G, De Paoli P. Heterogeneity of cag genotypes in Helicobacter pylori isolates from human biopsy specimens. J Clin Microbiol. 2003;41:976-980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 46. | Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Labo-ratory Manual. 2nd ed. New York: Cold Spring Harbor Labora-tory Press 1989; 1.21-1.52, 2.60-2.80, 7.3-7.35, 9.14-9.22. |
| 47. | Chen Y, Wang J, Shi L. [In vitro study of the biological activities and immunogenicity of recombinant adhesin of Heliobacter pylori rHpaA]. Zhonghua Yi Xue Zazhi. 2001;81:276-279. [PubMed] |
| 48. | Scholte GH, van Doorn LJ, Cats A, Bloemena E, Lindeman J, Quint WG, Meuwissen SG, Kuipers EJ. Genotyping of Helicobacter pylori in paraffin-embedded gastric biopsy specimens: relation to histological parameters and effects on therapy. Am J Gastroenterol. 2002;97:1687-1695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 49. | Kim JM, Kim JS, Jung HC, Oh YK, Kim N, Song IS. Inhibition of Helicobacter pylori-induced nuclear factor-kappa B activation and interleukin-8 gene expression by ecabet sodium in gastric epithelial cells. Helicobacter. 2003;8:542-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 50. | Machado JC, Figueiredo C, Canedo P, Pharoah P, Carvalho R, Nabais S, Castro Alves C, Campos ML, Van Doorn LJ, Caldas C. A proinflammatory genetic profile increases the risk for chronic atrophic gastritis and gastric carcinoma. Gastroenterology. 2003;125:364-371. [RCA] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 361] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 51. | Beales IL, Crabtree JE, Scunes D, Covacci A, Calam J. Antibodies to CagA protein are associated with gastric atrophy in Helicobacter pylori infection. Eur J Gastroenterol Hepatol. 1996;8:645-649. [PubMed] |
| 52. | Hoshino FB, Katayama K, Watanabe K, Takahashi S, Uchimura H, Ando T. Heterogeneity found in the cagA gene of Helicobacter pylori from Japanese and non-Japanese isolates. J Gastroenterol. 2000;35:890-897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
Edited by Chen WW and Wang XL Proofread by Xu FM