INTRODUCTION
In recent years it has become clear that angiogenesis is not only important in physiological processes such as embryonic development, wound healing, and organ and tissue regeneration, but also plays a pivotal role in tumor progression and metastasis[1]. Neovascularization is critical for the rapid growth of solid tumors[2]. However, most if not all solid tumors appear to be dependent on angiogenesis for their further expansion and preventing tumor cell apoptosis when their size exceeds beyond 1-2 cubic millimeters[3]. Tumor cells promote angiogenesis by secreting angiogenic factors, especially vascular endothelial growth factor (VEGF) or vascular permeability factor (PVF)[4].
VEGF is a specific mitogen for endothelial cells that has been shown to be expressed in many tumor cell lines and vascular smooth muscle cells[5,6] and is important not only for angiogenesis, but also for maintenance of existing blood vessels[7]. VEGF characterized by rapidly growing solid tumors can also derive from immune cells that infiltrate tumors[8]. Increased serum VEGF is found in cancer patients[9] and elevated VEGF levels are reported to be a prognostic clinical factor correlated with decreased survival in patients with breast, ovarian, lung, gastric, liver and colon[10,11] cancers.
Inhibition of VEGF action that controls tumor-induced angiogenesis on a molecular basis may therefore be a successful approach to the treatment of human hepatocarcinomas. Since VEGF is also known as the vascular permeability factor (VPF)[12], inhibition of VEGF may induce a dual therapeutic effect (e.g., antiangiogenic and anti-edematous) in patients with hepatocarcinoma[13].
We constructed successfully antisense VEGF165 eukaryotic expression vector PCDNA3-as-VEGF165 and transferred it into human hepatocarcinoma cells in vitro with cation lipofectamine 2000 mediated methods to evaluate the expression of VEGF protein and the inhibitory effect on the proliferation of hepatocarcinoma cells.
MATERIALS AND METHODS
Plasmid PCDNA3-as-VEGF165 constructions
The SMMC-7721 cell line had a high expression of endogenous VEGF protein, and used in antisense VEGF165 transfection studies. The entire coding region of human VEGF165 cDNA (0.6 kb) containing the 165 amino acid coding regions, was obtained from the PSV-VEGF165 plasmid (Preserved by professor Bai-Gen Zhang, Renji Hospital, Shanghai, China), and subcloned into the Pbluescript M13- expression vector (preserved by Biochemistry Department of Nanjing University, Nanjing, China) generating the Pbluescript –VEGF165 plasmid with endonucleases EcoR I and Pst I (MBI) digested. Then the Pbluescript –VEGF165 was digested by EcoR I and BamH I to obtain VEGF165 cDNA that was reverse inserted into the PCDNA3 eukaryon expression vector (preserved by Shanghai Institute for Biologic Science, Shanghai, China) generating the antisense VEGF165 plasmid PCDNA3-as-VEGF165. It was confirmed by restriction endonuclease analysis, PCR (Roche) and nucleotide sequencing (Shanghai Sangon Biological Engineering Technology and Service Co. Ltd., China). Then the antisense VEGF165 plasmid was amplified by transferring into activated Ecol. DH5α was extracted with QIAGEN plasmid purification kit (QIAGEN) following the manufacturer’s instructions.
ELISA for VEGF protein in cell culture supernatant
The day before transfection, the SMMC-7721 cells were trypsinized, counted, and plated in 24-well plates at 1 × 105 cells per well so that they were 90% - 95% confluent on the day of transfection. Cells were plated in 0.5 ml of DMEM containing 10% fetal bovine serum (FBS) and no antibiotics. For each well of cells to be transfected, 1.0 μg of DNA (PCDNA3 or PCDNA3-as-VEGF165) and 3.0 μl of lipofectamine 2000 (LF2000) reagent were diluted respectively into 50 μl of DMEM without serum and antibiotics, and incubated for 5 min at room temperature. Once the LF2000 reagent was diluted, it was combined with DNA within 30 min, and then incubated at room temperature for 20 min to allow DNA-LF2000 reagent complexes to form. The DNA-LF2000 reagent complexes (100 μl) were added directly to each well and mixed gently by rocking the plate back and forth. The cells were incubated at 37 °C in a CO2 incubator for 48 h until they were ready to assay for transgene expression. The amount of human VEGF protein in cell culture supernatant was quantified by using an ELISA kit for human VEGF (Quantikine, R and D Systems)[14].
Semiquantitative reverse transcription (RT)-PCR analysis[15]
Tripure RNA isolation system (Roche) was used to isolate RNA from hepatocarcinoma SMMC-7721 cells after the transfection of DNA-LF2000 reagent complexes. According to the manufacturer’s instructions, total RNA was resuspended in nuclease-free water (Promega), and the absorbance was read at A260 and A280. The quality of RNA was checked on a 1% denaturing agarose gel to ensure the presence of 28 S and 18 S ribosomal bands. VEGF mRNA expressions were measured using semiquantitative RT-PCR. One μg of total RNA was added to a 50 μl RT-PCR reaction (PCR-Access, Promega). The reaction master mix was prepared according to the manufacturer’s instructions with β-actin (as an internal control for both qualitative and semiquantitative RT-PCR) and VEGF gene-specific primers. The primer sequences were 5_-GAGGGCAGAATCATCACGAAGT -3_ (sense) and 5_-TCCTATGTGCTGGCCTTGGTGA -3_ (antisense) for VEGF. The primer sequences for β-actin amplification were 5_- CCAAGGCCAACCGCGAGAAGATGAC -3_ (sense) and 5 _ - AGGGTACATGGTGGTGCCGCCAGAC _ (antisense). For quantitation of mRNA, primers were used in a reaction involving one cycle of reverse transcription at 48 °C for 45 min and 30 cycles of denaturation at 94° for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 45 s. Then the resulting RT-PCR fragments were electrophoresed on 2% agarose gels at 110 V for 35 min.
VEGF165 cDNA PCR detection
Highpure DNA isolation system (Roche) was used to isolate DNA from normally cultured hepatocarcinoma SMMC-7721 cells. The primer sequences were 5_- GAGGGCAGAATC ATCACGAAGT -3_ (sense) and 5_- TCCTATGTGCTGGCCTTGGTGA -3_ (antisense) for VEGF. VEGF PCR was 30 cycles of denaturation at 94 °C for 1min, annealing at 55 °C for 1 min, and extension at 72 °C for 45 s. Then the fragments were identified by electrophoresis.
Flow cytometric analysis
The survival rate and apoptotic rate of hepatocarcinoma SMMC-7721 cells after antisense VEGF165 transfections were determined by propidium iodide (PI) and annexin V staining. Hepatocarcinoma cells were digested by 0.25% trypsin at 37 °C for 20 min and fixed with ice-cold 70% ethanol at a cell density of 1 × 106 ml-1. PI and annexin V were then added and incubated with cells in the dark for 30 min until detected by flow cytometry[16].
MTT assay
The day before PCDNA3-as-VEGF165 transfection, the hepatocarcinoma SMMC-7721 cells were trypsinized and counted, plated in 96-well plates at 1 × 104cells per well. Cells were plated in 100 μl of DMEM containing serum and no antibiotics, and incubated at 37 °C in a CO2 incubator for 48 h until they were ready to transfer antisense VEGF165. For each well of cells to be transfected, 0.2 μg of DNA (PCDNA3 or PCDNA3-as-VEGF165) and 0.6 μl LF2000 were diluted respectively into 50 μl of DMEM without serum. The DNA-LF2000 reagent complexes (100 μl) were added directly to each well and mixed gently by rocking the plate back and forth. The cells were incubated at 37 °C in a CO2 incubator for 48 h until they were ready to assay for proliferation of hepatocarcinoma cells by MTT.
MTT assay was performed according to the methods of Mosmann[17]. After the culture supernatant was sucked out, 100 μl 5 mg·ml-1 MTT (Sigma) stock solution in PBS was respectively added to 4 wells each group each day for 5 days. After 4 h of incubation, 100 μl DMSO (Sigma) containing serum was added into each well. The plate was mixed gently by rocking back and forth until the blue sedimentation was completely dissolved. Then the absorbances were read by Tecan’s sunrise absorbance microplate reader (A-5082)[18].
Immunohistochemistry[19]
The gene transferred hepatocarcinoma SMMC-7721 cells were trypsinized and sliced by cytofuge (Cytocentrifuge System M801-22, StatSpin). The slices containing SMMC-7721 cells were dried at 37 °C overnight, rehydrated and exposed to 0.5 per cent hydrogen peroxide for 30 min to block the endogenous peroxidase activity. They were washed with distilled water and TRIS-HCL buffered saline (TBS) at pH7.6, and then incubated with 1:5 normal rabbit serum for 10 min to reduce background staining. Next, the slices were incubated with diluted primary antibody at room temperature for 1 h. The antibody, a mouse monoclonal anti-human VEGF (DAKO, UK) was applied at a dilution of 1:200. The slices were then washed and a secondary antibody, horseradish peroxidase-conjugated rabbit anti-mouse immunoglobulin (DAKO, UK) at 1:50 dilution was applied for 30 min at room temperature. Thereafter, the slices were reacted in hydrogen peroxide-diaminobenzidine (DAB) solution to produce a brown color and counterstained with haematoxylin.
TBS was used as washing buffer between each stage of the staining and TBS containing 1 per cent bovine serum albumin was used to dilute the antibody.
Statistical calculations
The data were expressed as mean ± SD. Besides a two-sided Student t test with a level of significance at 5%, a multiple-factor ANOVA (Tukey’s test) was performed to analyze the data of different groups. All the data were analyzed with the statistical software SPSS10.0.
RESULTS
Identification of eukaryotic expression vector PCDNA3-as-VEGF165
The eukaryotic expression vector PCDNA3-as-VEGF165 was identified by restriction endonucleases cut with EcoR I and BamH I and electrophoresis generating a 600 bp fragment according to the result from GeneBank. The fragment obtained from PCR was 258 bp in length. VEGF165 cDNA was reverse subcloned into the site after T7 promoter appeared on the plasmid PCDNA3 by nucleotide sequencing, coinciding completely with the sequence from GeneBank. There were no endonuclease cut sites of EcoR I, Pst I and BamH I by zymogram analysis.
Detection of SMMC 7721 cells after PCDNA3-as-VEGF165 transfection
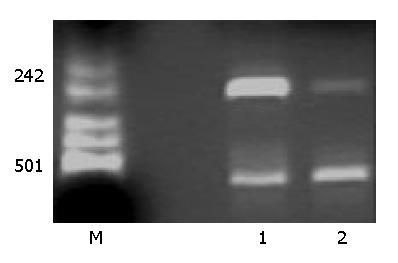
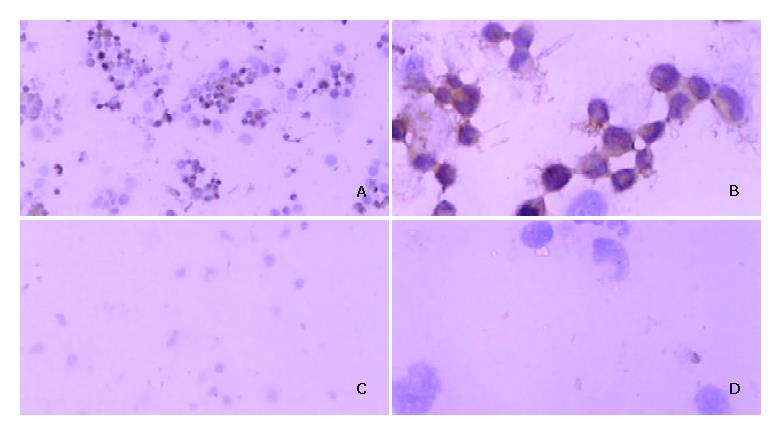
VEGF cDNA was highly expressed in normally cultured hepatocarcinoma SMMC-7721 cells by PCR. But the VEGF mRNA expression was dramatically decreased after PCDNA3-as-VEGF165 transfection in hepatocarcinoma SMMC-7721 cells (Figure 1). The expression of VEGF protein in the culture supernatant of hepatocarcinoma SMMC-7721 cells was significantly inhibited (142.01 ± 7.95 vs 1 625.52 ± 64.46 pg·ml-1, P < 0.01) 2 days after antisense VEGF165 transfection when compared to PCDNA3 transfected controls, which correlated with the dose of antisense VEGF165 gene. VEGF protein was highly expressed in most normally cultured hepatocarcinoma cells by immunohistochemical staining, but only in few hepatocarcinoma cells 2 days after antisense VEGF165 transfection (Figure 2).
Figure 1 Effect of PCDNA3-as-VEGF165 on VEGF mRNA ex-pression of hepatocarcinoma SMMC-7721 cells by RT-PCR.
VEGF mRNA expression was negative after PCDNA3-as-VEGF165/LF2000 transfected. (Lane 1: PCDNA3/LF2000 transfected, Lane 2: PCDNA3-as-VEGF165/LF2000 transfected; β-actin: 587 bp, VEGF 258 bp).
Figure 2 Immunohistochemical staining for expression of VEGF protein after DNA/LF2000 transfection in hepatocarcinoma SMMC-7721 cells.
Positive signal for VEGF protein is brown and located in cytoplasm. A: (×100), B: (×400): PCDNA3 transferred SMMC-7721 cells; C: (×100), D: (×400): PCDNA3-as-VEGF165 transferred.
Evaluation of functions of eukaryotic expression vector PCDNA3-as-VEGF165
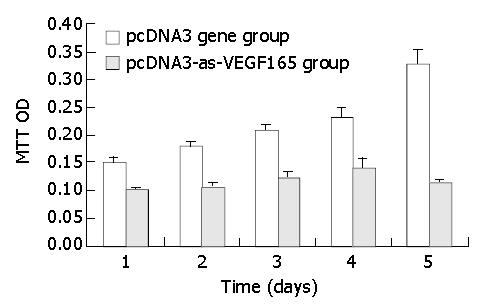
The apoptotic rate of hepatocarcinoma cells was significantly promoted (17.98% ± 0.86% vs 4.86% ± 0.27%, P < 0.01) and the survival rate was notably decreased (80.99% ± 3.20% vs 93.52% ± 3.93%, P < 0.05) due to antisense VEGF165 by flow cytometry (FCM). The transfection of antisense VEGF165 gene resulted in the inhibition on the proliferation of hepatocarcinoma cells using MTT and the death of all hepatocarcinoma cells on day 6 after transfection (Figure 3).
Figure 3 Effect of PCDNA3-as-VEGF165 on proliferation of hepatocarcinoma SMMC-7721 cells by MTT.
DISCUSSION
The incidence of hepatic cancer has been gradually increasing in recent years in China. Currently surgical operation is considered exclusively as the sole option for the treatment of hepatocarcinoma, although less than 20% of patients were considered as candidates for resection [20]. hepatocarcinoma is clearly a disease for which alternative therapies must be developed.
VEGF-mediated angiogenesis was considered to be a major contributor to tumor growth and metastasis[21,22]. A number of VEGF antagonists have been developed for inhibiting VEGF-induced tumor angiogenesis and tumor growth. These antagonists include antisense VEGF oligonucleotides, VEGF antibodies, dominant-negative receptors, and soluble VEGF receptors [17,23-25].
VEGF has been found to band to two tyrosine kinase receptors, VEGFR-1 (Flt-1) and VEGFR-2 (KDR/ Flk-1)[26-28]. Both receptors were expressed on the surface of tumor endothelial cells, but not on tumor cells[29]. Antisense VEGF cDNA[30,31] significantly inhibited tumor angiogenesis and tumor growth in vivo with a paracrine signal transduction system. Angiogenesis played a key role in tumor growth and metastasis.
In previous studies, the putative anti-angiogenic effect of antisense VEGF165 was investigated on stably transfected tumor cell lines. C6 and U87 glioma cells stably transfected with a vector encoding antisense VEGF165 showed a significant growth inhibition when compared to sense VEGF transferred controls[32,33]. These studies suggested that antisense VEGF165 expression could be useful in controlling tumor angiogenesis and tumor growth in vivo.
In order to increase the effect of putative gene therapy, we implanted PCDNA3-as-VEGF165/LF2000 complexes into human hepatocarcinoma SMMC-7721 cells. We used hepatocarcinoma cells, because we previously observed that normally cultured hepatocarcinoma SMMC-7721 cells showed a high expression of VEGF protein[34]. We achieved a significant promotion of SMMC-7721 cell apoptosis and complete hepatocarcinma cell death on day 6 after antisense VEGF165 transfection. VEGF mRNA expression was obviously blocked by antisense VEGF165. Since the effect of antisense VEGF mRNA expression was temporary, it bound specifically to the sense counterpart and selectively neutralized the function of VEGF165 gene[35]. This approach may also require persistent expression of antisense VEGF or periodic administration of PCDNA3-as-VEGF165.
In summary, we succeeded in constructing the antisense VEGF165 eukaryotic expression vector PCDNA3-as-VEGF165 acting as a VEGF165 antagonist, and studied its anti-tumor activity. But the molecular mechanisms of PCDNA3-as-VEGF165 mediated apoptosis and inhibitory proliferation of hepatocarcinoma cells still need to be established. Finally, there may also exist other angiogenic pathways in hepatocarcinoma such as the angiopoietin/tie2 system that are not blocked by antisense VEGF165[36].