Published online Feb 1, 2004. doi: 10.3748/wjg.v10.i3.371
Revised: August 9, 2003
Accepted: August 16, 2003
Published online: February 1, 2004
AIM: To study the nuclear microsatellite instability (nMSI) at BAT26 and mitochondral microsalellite instability (mtMSI) in the occurrence and development of hepatocellular carcinoma and the relationship between nMSI and mtMSI.
METHODS: nMSI was observed with PCR and mtMSI with PCR-SSCP in 52 cases of hepatocellular carcinoma.
RESULTS: mtMSI was detected in 11 out of the 52 cases of hepatocellular carcinoma (21.2%). Among the 11 cases of hepatocellular carcinoma with mtMSI, 7 occured in one locus and 4 in 2 loci. The frequency of mtMSI in the 52 cases of hepatocellular carcinoma showed no correlation to sex, age, infection of hepatitis B, liver cirrhosis as well as positive AFP of the patients (P > 0.05). In addition, nMSI was detected in 3 out of 52 cases of hepatocellular carcinoma (5.8%) and there was no correlation of the incidence of mtMSI to that of nMSI (P > 0.05).
CONCLUSION: mtMSI may be involved in the coccurrence and development of hepatocellular carcinoma and it is independent of nMSI.
- Citation: Fang DC, Fang L, Wang RQ, Yang SM. Nuclear and mitochondrial DNA microsatellite instability in Chinese hepatocellular carcinoma. World J Gastroenterol 2004; 10(3): 371-375
- URL: https://www.wjgnet.com/1007-9327/full/v10/i3/371.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i3.371
Mitochondria are the energy-transducting organelles of eukaryotic cells in which fuels to drive cellular metabolism are converted into cellular adenosine triphosphate (ATP) through the process of oxidative phosphorylation. Mitochondria are responsible for generating approximately 90% of ATP. The mitochondrion is the only organelle in the cell, aside from the nucleus, which contains its own genome and genetic machinery[1]. Mitochondrial DNA (mtDNA) is a 16569 base-pair, double-stranded and closed circular molecule, and encodes 13 polypeptides. All of the polypeptides are components of the respiratory chain/OXPHOS system, plus 24 genes, specifying two ribosomal RNAs (rRNAs) and 22 transfer RNAs (tRNAs), which are required to synthesize the 13 polypeptides. Mitochondrial genome is far more vulnerable to oxidative damage and undergoes a higher rate of mutation than nuclear genome due to its lack of histone protection, limited repair capacity, and close proximity to the electron transport chain, which constantly generates superoxide radicals[2-5]. Accumulation of mutations in mtDNA is approximately tenfold greater than that in nuclear DNA[6].
A high frequency of mtDNA mutations has been identified in cancer of the colon[7], stomach[8], liver[9], pancreas[10], lung[11], breast[12], kidney[13], prostate[14], ovary[15], Barrett’s esophagus[16] and leukemia[17]. The majority of these somatic mutations were homoplasmic, suggesting that mutant mtDNA becomes dominant in tumor cells. In addition, microsatellite instability has also been shown in mtDNA of colorectal and gastric carcinomas[18,19] . Further studies demonstrated that mononucleotide could repeat alteration, missense mutation, and small deletion in NADH dehydrogenase genes and alteration in a polycytidine (C)n tract in the D-loop region of mtDNA could occur in colorectal carcinomas[20]. These results imply that microsatellite instability in mtDNA (mtMSI) of colorectal carcinoma may be resulted from certain deficiencies in DNA repair. Therefore, it has been proposed that somatic mutations and mtMSI play a role in tumorigenesis and development of cancer[21].
Hepatocellular carcinoma (HCC) is one of the most common causes of cancer related mortality worldwide. The incidence of HCC shows a considerable geographical variation with a very high incidence in China. Epidemiological studies in high-risk populations have identified chronic hepatitis B virus (HBV) and chronic hepatitis C virus (HCV) infection as well as dietary exposure to aflatoxin B1 (AFB1) as major factors in the etiology of this disease[22]. It has been reported that the amount of AFB1 combined to hepatocellular mtDNA is 3 - 4 fold larger than that combined to nuclear DNA(nDNA). This combined product of aflatoxin cannot easily be expelled and stays in mtDNA for a long period[22]. Since there is a prolonged period between initial HBV and HCV infection and emergence of HCC, multiple genetic events may occur to promote the malignant transformation of hepatocytes. Many chromosomal aberrations have been frequently reported in HCCs including loss of heterozygosity (LOH) at numerous loci[23,24]. The repeated destruction and regeneration of liver tissue associated with chronic viral hepatitis would lead to accumulation of mtDNA mutations[25]. Although MSI in nuclear DNA (nDNA) of HCCs has been detected[26-32], little attention has been paid to MSI in mtDNA(mtMSI) in this tumor. In order to elucidate the role of mtMSI in the hepatocarcinogenesis, we examined mtMSI and nMSI in a set of 52 Chinese HCCs.
Fresh tissues were collected from 52 HCC patients undergoing hepatic resection in the Southwest Hospital, Third Military Medical University, Chongqing, China from 1996 to 2002. Neoplastic and nonneoplastic liver tissues were frozen in liquid nitrogen immediately and kept at -70 °C until processing. The 52 patients consisted of 42 males and 10 females, their age ranged from 22 to 71 years with an average of 48.8 years at diagnosis. Thirty-two patients were positive and 20 were negative for hepatitis B surface antigen (HBsAg). Hepatitis C virus antibody (Anti-HCV) was negative for all cases. Hematoxylin and eosin-stained sections were prepared from the same samples used for mtMSI and nMSI studies and the diagnosis of HCC was confirmed by histology. None of the patients included in the present series had a family history suggestive of HNPCC and none had received previous chemotherapy or radiation therapy. Necrotic tumors were excluded from the study. The tumor samples contained more than 70% malignant cells. Genomic DNA was isolated from tumor and non-tumor liver tissues and blood, using standard proteinase-K digestion and phenol-chloroform extraction protocols.
PCR-single strand conformation polymophism (PCR-SSCP) was performed to amplify the microsatellite sequence of mtDNA using published primers[18]. The primer consisted of 2 D-loop regions and 5 coding regions (Table 1). The reaction conditions and procedures were similar to those reported by Hebano et al[18].
| Repeat sequence | mtDNA region | Position | Annealing(°C) | Primer (5’-3’) |
| (C)n | 270 - 425 | D - loop | 58 | TCCACACAGACATCAATAACA |
| AAAGTGCATACCGCCAAAAG | ||||
| (CA)n | 467 - 556 | D - loop | 55 | CCCATACTACTAATCTCATCAA |
| TTTGGTTGGTTCGGGGTATG | ||||
| (C)6 | 3529 - 3617 | ND1 | 55 | CCGACCTTAGCTCTCACCAT |
| AATAGGAGGCCTAGGTTGAG | ||||
| (A)7 | 4555 - 4644 | ND2 | 55 | CCTGAGTAGGCCTAGAAATAAA |
| ACTTGATGGCAGCTTCTGTG | ||||
| (T)7 | 9431 - 9526 | COIII | 55 | CCAAAAAGGCCTTCGATACG |
| GCTAGGCTGGAGTGGTAAAA | ||||
| (C)6 and (A)8 | 12360 - 12465 | ND5 | 55 | CACCCTAACCCTGACTTCC |
| GGTGGATGCGACAATGGATT | ||||
| (CCT)3 and (AGC)3 | 12940 - 13032 | ND5 | 55 | GCCCTTCTAAACGCTAATCC |
| TCAGGGGTGGAGACCTAATT |
Each PCR was digested by appropriate restriction enzymes and electrophoresed at 300V at 22 °C for 2 hr on a 7.5% polyacrylamide gel containing 50 mmol/L boric acid, 1 mmol/L EDTA and 2.5% glycerol. After silver staining, PCR products showing mobility shifts were directly sequenced using appropriate internal primer and analyzed using 373A automated DNA sequencer (Perkin Elmer Cetus). All analyses were performed twice to rule out PCR artifact.
MSI at BAT26 microsatellite locus was analyzed using PCR method. The sequence of upper stream primer was 5’-TGACTACTTTTGACTTCAGCC-3’ and that of down stream primer was 5’-AAC CAT TCA ACA TTT TTA ACC C-3’. PCR was performed in 20 ml of reaction mixture containing 10 mmol/L Tris-HCl (pH8.3), 50 mmol/L KCl, 1.5 mmol/L MgCl2, 200 mmol/L each deoxyncleotide triphoshate, 0.5 mmol/L of each primer, 0.5 unit Ampli Taq polymerase (Perkin-Elmer Cetus, NowalK), 100 ng genomic DNA and 0.5 mCi [33p] dATP. The reaction was carried out in a thermal cycler at 94 °C for 1 min, at 55 °C - 62 °C for 1 min, and at 72 °C for 1min, for 35 cycles with an initial denaturation step at 94 °C for 5 min and final extension step at 72 °C for 10 min. The PCR products were then separated on 5% polyacrylamide 7M urea denaturing gel, and visulized by autoradiography. MSI was defined as the presence of a band shift in the tumor DNA not present in the corresponding normal DNA.
χ2 test was used for statistical analysis and P < 0.05 was considered as statistically significant.
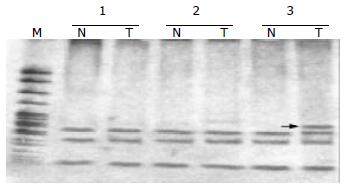
Fifty-two HCC samples were screened for mtMSI at seven repeat sites using the PCR-RFLP method. Figure 1 exhibits a representative mobility-shift band compared with normal counterpart. mtMSI affecting at least one locus was observed in 11 out of 52 cases (21.2%), in which 7 cases affected 1 locus and 4 cases affected 2 loci. mtMSI occurred in D-loop in 10 cases (19.2%), in which 8 cases occurred in (C)n region and 2 cases in (CA)n region. mtMSI occurred in the coding region in 5 cases (9.6%), and concomitant mtMSI locus was found in the D-loop in 4 out of the 5 cases. The frequency of mtMSI in 52 cases of HCC showed no correlation to sex, age, HBV infection, liver cirrhosis and positive AFP of the patients (P > 0.05, Table 2).
| n | mtMSI positive | mtMSI negative | ||
| Sex | Male | 42 | 10 | 32 |
| Female | 10 | 1 | 9 | |
| Age | < 30 | 1 | 1 | 0 |
| 30 - 60 | 41 | 8 | 33 | |
| ≥ 60 | 10 | 2 | 8 | |
| HBsAg | Positive | 38 | 8 | 30 |
| Negative | 14 | 3 | 11 | |
| Cirrhosis | Positive | 37 | 9 | 28 |
| Negative | 15 | 2 | 13 | |
| AFP | Positive | 26 | 6 | 20 |
| Negative | 26 | 5 | 21 |
The mobility shift in tumor DNA compared to corresponding normal DNA samples representing nMSI is shown in Figure 2. nMSI was found in 3 of 52 cases of HCC (5.8%). In the 3 cases of nMSI, only 1 case showed mtMSI simultaneously. No correlation was found between nMSI and mtMSI in the 52 cases of HCC.
It has been discovered so far that mitochondria are the only organelle to have their own genome and to undergo replication, transcription and translation without dependence on nuclear DNA. They are called as the “25th chromosome of human body”. Many diseases have been found to be related to the structural and functional defects of mitochondria and consequently they are known as mitochondrial diseases[33]. mtMSI has been found to be a very common phenomenon accompanying gastric carcinoma, colorectal carcinoma and breast carcinoma and may play an important role in the carcinogenesis of these malignant diseases[12,18-21]. To study the role of mtMSI in liver carcinogenesis, we analyzed 52 cases of HCC using seven microsatellite markers known to be altered in gastrointestinal carcinomas. mtMSI in at least one locus was found in 11 of the 52 cases (21.2%) of HCC, implying that mtMSI might occur not only in gastrointestinal cancers but also in hepatic cancers, and it may play an important role in the occurrence of a certain number of HCC.
Unlike other types of cancer, HCC has been found usually preceded by chronic inflammation due to viral infection[34-37]. Matsuyama et al[11] reported that the frequency of mtDNA mutations was markedly increased in both noncancerous and cancerous liver specimens compared with control liver tissue. Accumulation of mtDNA mutations in HCC tissue could reflect its malignant potency. The frequency of mtDNA mutations was significant higher in HBV infection-related HCC than in other tumors, which implies that repeated destruction and regeneration of the liver tissue associated with chronic viral hepatitis would lead to accumulation of mtDNA mutations. In the current study, we did not find any obvious relationship between mtMSI and HBsAg, suggesting that HBV infection might play a limited role in the mtMSI pathway of HCC. In addition, we did not find an obvious relationship between mtMSI and sex, age, cirrhosis as well as positive AFP.
MtDNA contains several mono- and dinucleotide repeats. The most frequently used mtDNA in the test of mtMSI is a (CA)n microsatellite starting at 514 bp position of the D-loop[38] and a homopolymeric C tract extending from 16184 to 16193 bp of the D-loop, which could be interrupted by a T at 16189 bp position[39]. Alonso et al[40] studied mutations in the mtDNA D-loop region and found three mutations in eight gastric tumors. Richard et al[41] studied 40 pairs of normal/ cancer breast specimens for the presence of mtMSI and found a 216-fold increase in the D-loop point mutations of cancer cells with regard to the spontaneous rate detected in female gametes. Maximo et al[19] utilized PCR-SSCP to examine mtDNA large deletions and mutations in 32 gastric carcinomas and found that most of the mutations corresponded to insertions/deletions in the D-loop region or transitions in ND1, ND5, and COXI. Earlier studies revealed the presence of mutations in the D-loop of both non-malignant and malignant gastric tumors[42,43]. Analysis of HCCs indicated that mutations in the D-loop were a frequent event and could be used as a molecular tool for the determination of clonality[9,44]. Two recent studies reported the frequency of D-loop mutations in esophageal cancer. One group focused on adenocarcinomas of Barrett’s esophagus. In that study, D-loop alterations were identified in 40% of the patients examined[16]. The other study showed that D-loop mutations were much less frequent in esophageal cancer, occurring in only 5% of the specimens analyzed[45]. Clearly, analysis of mtDNA from more esophageal tumor samples is needed in order to determine the frequency of D-loop mutations and their relevance in this type of cancer. In our series of 52 cases of HCC, mtMSI was found in 11 (21.2%). MtMSI occurred in the D-loop region of 10 cases and in the coding region of 5 cases. Among the 10 cases, mtMSI occurred in (C)n region of 8 cases and in (CA)n region of 2 cases, So (C)n region of D-loop is the site at which mtMSI occurs more frequently than in other regions. Our findings are consistent to those reported by Habano et al[18-20] and Maximo et al[18-20].
Microsatellite markers might provide evidences of faulty DNA mismatch repair (MMR) via the detection of MSI[46-49]. The choice of microsatellite markers may impact on the MSI detection rate. BAT-26, a repeat of 26 deoxyadenosine localized in intron 5 of hMSH2 gene, has been reported as a reliable indicator of replication error phenotype in colorectal cancers, enabling analysis of tumour DNA in the absence of paired normal DNA[50]. The frequency of nMSI in hepatic cancer varied in different reports[51,52]. Karachristos et al[52] studied 27 cases of HCC and found none of the tumors examined showed alterations in BAT-26. In our series of 52 cases, 3 cases were found to have nMSI at BAT26 (5.8%).Our finding indicates that nMSI at BAT26 is not common in cases of HCC and support the hypothesis that HCC is a “low” MSI tumor in China. Carcinogenesis of HCC may undergo a different molecular route other than that of nMSI.
Mutation of mtDNA may result in the occurrence of tumor but its mechanism remains unknown. Further studies are required to determine if mtDNA mutations are correlated with malignant transformation. Recently, scholars have shifted their attention to the interactions between mtDNA and nDNA. Fragments of mtDNA are sometimes found in nuclear genes, and the insertion of mtDNA has been suggested as a mechanism by which oncogenes are activated[53]. For example, sequences representing subunits ND4 (Complex I) and subunits cytochrome C oxidases I, II and III (complex IV) have been found in the nuclear DNA of various tissues[53]. In yeast cells, migration of DNA from the mitochondria to the nucleus occurred 100000 times more frequently than in the opposite direction[54]. In our series of 52 cases of HCC, nMSI was detected in 5.8% and coexistance of nMSI and mtMSI in only 1 out of 3 cases. We failed to confirm there was a correlation of mtMSI to nMSI in our cases of HCC. This finding is in agreement with the recently published data on gastrointestinal cancer[55].
In conclusion, mtMSI could play an important role at multiple stages in the process of carcinogenesis. The mitochondrial production of ROS might be involved in the initiation and promotion of carcinogenesis, in part due to ROS-triggered mutagenesis of both mtDNA and nDNA[56]. Also, other evidences exists for a mechanism of nDNA mutagenesis involving the integration of mtDNA fragments. Many primary tumors revealed a high frequency of mtDNA mutations and the majority of these somatic mutations were homoplasmic in nature, indicating that the mutant mtDNA has become dominant in tumor cells. The mutated mtDNA was readily detectable in paired bodily fluids from each type of cancer and was 19 to 220 times as abundant as mutated nuclear p53 DNA. By virtue of their clonal nature and high copy number, mitochondrial mutations might provide a powerful molecular marker for noninvasive detection of cancer[57]. Important areas for future research should include intergenomic signaling pathways in carcinogenesis and the potential role of mitochondria and mtDNA mutations in immunological surveillance of tumor cells. Finally, the role of mitochondria in stimulating apoptosis could be exploited in cancer therapeutics[58].
| 1. | Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6507] [Cited by in RCA: 6541] [Article Influence: 145.4] [Reference Citation Analysis (0)] |
| 2. | Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA. 1997;94:514-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1282] [Cited by in RCA: 1380] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 3. | Marcelino LA, Thilly WG. Mitochondrial mutagenesis in human cells and tissues. Mutat Res. 1999;434:177-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 101] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 4. | Kunkel TA, Loeb LA. Fidelity of mammalian DNA polymerases. Science. 1981;213:765-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 131] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 5. | Shay JW, Werbin H. Are mitochondrial DNA mutations involved in the carcinogenic process. Mutat Res. 1987;186:149-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 100] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 6. | Johns DR. Seminars in medicine of the Beth Israel Hospital, Boston. Mitochondrial DNA and disease. N Engl J Med. 1995;333:638-644. [PubMed] |
| 7. | Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20:291-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 625] [Cited by in RCA: 621] [Article Influence: 22.2] [Reference Citation Analysis (5)] |
| 8. | Habano W, Sugai T, Nakamura SI, Uesugi N, Yoshida T, Sasou S. Microsatellite instability and mutation of mitochondrial and nuclear DNA in gastric carcinoma. Gastroenterology. 2000;118:835-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 97] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 9. | Nomoto S, Yamashita K, Koshikawa K, Nakao A, Sidransky D. Mitochondrial D-loop mutations as clonal markers in multicentric hepatocellular carcinoma and plasma. Clin Cancer Res. 2002;8:481-487. [PubMed] |
| 10. | Jones JB, Song JJ, Hempen PM, Parmigiani G, Hruban RH, Kern SE. Detection of mitochondrial DNA mutations in pancreatic cancer offers a "mass"-ive advantage over detection of nuclear DNA mutations. Cancer Res. 2001;61:1299-1304. [PubMed] |
| 11. | Matsuyama W, Nakagawa M, Wakimoto J, Hirotsu Y, Kawabata M, Osame M. Mitochondrial DNA mutation correlates with stage progression and prognosis in non-small cell lung cancer. Hum Mutat. 2003;21:441-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 12. | Tan DJ, Bai RK, Wong LJ. Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Cancer Res. 2002;62:972-976. [PubMed] |
| 13. | Nagy A, Wilhelm M, Kovacs G. Mutations of mtDNA in renal cell tumours arising in end-stage renal disease. J Pathol. 2003;199:237-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 14. | Jerónimo C, Nomoto S, Caballero OL, Usadel H, Henrique R, Varzim G, Oliveira J, Lopes C, Fliss MS, Sidransky D. Mitochondrial mutations in early stage prostate cancer and bodily fluids. Oncogene. 2001;20:5195-5198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 174] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 15. | Liu VW, Shi HH, Cheung AN, Chiu PM, Leung TW, Nagley P, Wong LC, Ngan HY. High incidence of somatic mitochondrial DNA mutations in human ovarian carcinomas. Cancer Res. 2001;61:5998-6001. [PubMed] |
| 16. | Miyazono F, Schneider PM, Metzger R, Warnecke-Eberz U, Baldus SE, Dienes HP, Aikou T, Hoelscher AH. Mutations in the mitochondrial DNA D-Loop region occur frequently in adenocarcinoma in Barrett's esophagus. Oncogene. 2002;21:3780-3783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 17. | Ivanova R, Lepage V, Loste MN, Schächter F, Wijnen E, Busson M, Cayuela JM, Sigaux F, Charron D. Mitochondrial DNA sequence variation in human leukemic cells. Int J Cancer. 1998;76:495-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 18. | Habano W, Nakamura S, Sugai T. Microsatellite instability in the mitochondrial DNA of colorectal carcinomas: evidence for mismatch repair systems in mitochondrial genome. Oncogene. 1998;17:1931-1937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 124] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 19. | Máximo V, Soares P, Seruca R, Rocha AS, Castro P, Sobrinho-Simões M. Microsatellite instability, mitochondrial DNA large deletions, and mitochondrial DNA mutations in gastric carcinoma. Genes Chromosomes Cancer. 2001;32:136-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 94] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 20. | Habano W, Sugai T, Yoshida T, Nakamura S. Mitochondrial gene mutation, but not large-scale deletion, is a feature of colorectal carcinomas with mitochondrial microsatellite instability. Int J Cancer. 1999;83:625-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 21. | Hochhauser D. Relevance of mitochondrial DNA in cancer. Lancet. 2000;356:181-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 22. | Niranjan BG, Bhat NK, Avadhani NG. Preferential attack of mitochondrial DNA by aflatoxin B1 during hepatocarcinogenesis. Science. 1982;215:73-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 132] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Rashid A, Wang JS, Qian GS, Lu BX, Hamilton SR, Groopman JD. Genetic alterations in hepatocellular carcinomas: association between loss of chromosome 4q and p53 gene mutations. Br J Cancer. 1999;80:59-66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 60] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 24. | Okabe H, Ikai I, Matsuo K, Satoh S, Momoi H, Kamikawa T, Katsura N, Nishitai R, Takeyama O, Fukumoto M. Comprehensive allelotype study of hepatocellular carcinoma: potential differences in pathways to hepatocellular carcinoma between hepatitis B virus-positive and -negative tumors. Hepatology. 2000;31:1073-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 74] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 25. | Nishikawa M, Nishiguchi S, Shiomi S, Tamori A, Koh N, Takeda T, Kubo S, Hirohashi K, Kinoshita H, Sato E. Somatic mutation of mitochondrial DNA in cancerous and noncancerous liver tissue in individuals with hepatocellular carcinoma. Cancer Res. 2001;61:1843-1845. [PubMed] |
| 26. | Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis--A comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32:970-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 197] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 27. | Wang G, Zhao Y, Liu X, Wang L, Wu C, Zhang W, Liu W, Zhang P, Cong W, Zhu Y. Allelic loss and gain, but not genomic instability, as the major somatic mutation in primary hepatocellular carcinoma. Genes Chromosomes Cancer. 2001;31:221-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Salvucci M, Lemoine A, Saffroy R, Azoulay D, Lepère B, Gaillard S, Bismuth H, Reynès M, Debuire B. Microsatellite instability in European hepatocellular carcinoma. Oncogene. 1999;18:181-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 29. | Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Microsatellite instability associated with hepatocarcinogenesis. J Hepatol. 1999;31:529-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 30. | Takagi K, Esumi M, Takano S, Iwai S. Replication error frequencies in primary hepatocellular carcinoma: a comparison of solitary primary versus multiple primary cancers. Liver. 1998;18:272-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 31. | Kazachkov Y, Yoffe B, Khaoustov VI, Solomon H, Klintmalm GB, Tabor E. Microsatellite instability in human hepatocellular carcinoma: relationship to p53 abnormalities. Liver. 1998;18:156-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 32. | Macdonald GA, Greenson JK, Saito K, Cherian SP, Appelman HD, Boland CR. Microsatellite instability and loss of heterozygosity at DNA mismatch repair gene loci occurs during hepatic carcinogenesis. Hepatology. 1998;28:90-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 60] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 33. | Pulkes T, Hanna MG. Human mitochondrial DNA diseases. Adv Drug Deliv Rev. 2001;49:27-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 34. | Shen LJ, Zhang HX, Zhang ZJ, Li JY, Chen MQ, Yang WB, Huang R. Detection of HBV, PCNA and GST-pi in hepatocellular carcinoma and chronic liver diseases. World J Gastroenterol. 2003;9:459-462. [PubMed] |
| 35. | Tang ZY. Hepatocellular carcinoma--cause, treatment and metastasis. World J Gastroenterol. 2001;7:445-454. [PubMed] |
| 36. | Guo SP, Wang WL, Zhai YQ, Zhao YL. Expression of nuclear factor-kappa B in hepatocellular carcinoma and its relation with the X protein of hepatitis B virus. World J Gastroenterol. 2001;7:340-344. [PubMed] |
| 37. | Gao FG, Sun WS, Cao YL, Zhang LN, Song J, Li HF, Yan SK. HBx-DNA probe preparation and its application in study of hepatocarcinogenesis. World J Gastroenterol. 1998;4:320-322. [PubMed] |
| 38. | Szibor R, Michael M, Spitsyn VA, Plate I, Ginter EK, Krause D. Mitochondrial D-loop 3' (CA)n repeat polymorphism: optimization of analysis and population data. Electrophoresis. 1997;18:2857-2860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 39. | Bendall KE, Sykes BC. Length heteroplasmy in the first hypervariable segment of the human mtDNA control region. Am J Hum Genet. 1995;57:248-256. [PubMed] |
| 40. | Alonso A, Martin P, Albarran C, Aquilera B, Garcia O, Guzman A, Oliva H, Sancho M. Detection of somatic mutations in the mitochondrial DNA control region of colorectal and gastric tumors by heteroduplex and single-strand conformation analysis. Electrophoresis. 1997;18:682-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 84] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 41. | Richard SM, Bailliet G, Páez GL, Bianchi MS, Peltomäki P, Bianchi NO. Nuclear and mitochondrial genome instability in human breast cancer. Cancer Res. 2000;60:4231-4237. [PubMed] |
| 42. | Tamura G, Nishizuka S, Maesawa C, Suzuki Y, Iwaya T, Sakata K, Endoh Y, Motoyama T. Mutations in mitochondrial control region DNA in gastric tumours of Japanese patients. Eur J Cancer. 1999;35:316-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 80] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 43. | Burgart LJ, Zheng J, Shu Q, Strickler JG, Shibata D. Somatic mitochondrial mutation in gastric cancer. Am J Pathol. 1995;147:1105-1111. [PubMed] |
| 44. | Liu MR, Pan KF, Li ZF, Wang Y, Deng DJ, Zhang L, Lu YY. Rapid screening mitochondrial DNA mutation by using denaturing high-performance liquid chromatography. World J Gastroenterol. 2002;8:426-430. [PubMed] |
| 45. | Hibi K, Nakayama H, Yamazaki T, Takase T, Taguchi M, Kasai Y, Ito K, Akiyama S, Nakao A. Detection of mitochondrial DNA alterations in primary tumors and corresponding serum of colorectal cancer patients. Int J Cancer. 2001;94:429-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 59] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 46. | Fang DC, Wang RQ, Yang SM, Yang JM, Liu HF, Peng GY, Xiao TL, Luo YH. Mutation and methylation of hMLH1 in gastric carcinomas with microsatellite instability. World J Gastroenterol. 2003;9:655-659. [PubMed] |
| 47. | Fang DC, Luo YH, Yang SM, Li XA, Ling XL, Fang L. Mutation analysis of APC gene in gastric cancer with microsatellite instability. World J Gastroenterol. 2002;8:787-791. [PubMed] |
| 48. | Cai Q, Sun MH, Lu HF, Zhang TM, Mo SJ, Xu Y, Cai SJ, Zhu XZ, Shi DR. Clinicopathological and molecular genetic analysis of 4 typical Chinese HNPCC families. World J Gastroenterol. 2001;7:805-810. [PubMed] |
| 49. | Fang DC, Yang SM, Zhou XD, Wang DX, Luo YH. Telomere erosion is independent of microsatellite instability but related to loss of heterozygosity in gastric cancer. World J Gastroenterol. 2001;7:522-526. [PubMed] |
| 50. | Cravo M, Lage P, Albuquerque C, Chaves P, Claro I, Gomes T, Gaspar C, Fidalgo P, Soares J, Nobre-Leitão C. BAT-26 identifies sporadic colorectal cancers with mutator phenotype: a correlative study with clinico-pathological features and mutations in mismatch repair genes. J Pathol. 1999;188:252-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 51. | Martins C, Kedda MA, Kew MC. Characterization of six tumor suppressor genes and microsatellite instability in hepatocellular carcinoma in southern African blacks. World J Gastroenterol. 1999;5:470-476. [PubMed] |
| 52. | Karachristos A, Liloglou T, Field JK, Deligiorgi E, Kouskouni E, Spandidos DA. Microsatellite instability and p53 mutations in hepatocellular carcinoma. Mol Cell Biol Res Commun. 1999;2:155-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 53. | Corral M, Baffet G, Kitzis A, Paris B, Tichonicky L, Kruh J, Guguen-Guillouzo C, Defer N. DNA sequences homologous to mitochondrial genes in nuclei from normal rat tissues and from rat hepatoma cells. Biochem Biophys Res Commun. 1989;162:258-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 54. | Thorsness PE, Fox TD. Escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Nature. 1990;346:376-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 191] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 55. | Schwartz S, Perucho M. Somatic mutations in mitochondrial DNA do not associate with nuclear microsatellite instability in gastrointestinal cancer. Gastroenterology. 2000;119:1806-1808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 56. | Li JM, Cai Q, Zhou H, Xiao GX. Effects of hydrogen peroxide on mitochondrial gene expression of intestinal epithelial cells. World J Gastroenterol. 2002;8:1117-1122. [PubMed] |
| 57. | Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017-2019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 599] [Cited by in RCA: 612] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 58. | Shen ZY, Shen J, Li QS, Chen CY, Chen JY, Yi Z. Morphological and functional changes of mitochondria in apoptotic esophageal carcinoma cells induced by arsenic trioxide. World J Gastroenterol. 2002;8:31-35. [PubMed] |
Edited by Zhang JZ and Wang XL