Published online Dec 15, 2004. doi: 10.3748/wjg.v10.i24.3602
Revised: May 6, 2004
Accepted: May 13, 2004
Published online: December 15, 2004
AIM: Nucleocapsid (N) protein plays an important role in reproduction and pathological reaction of severe acute respiratory syndrome (SARS) coronavirus (SCoV), the antigenicity of the protein is better than spike (S) protein. This study was to find a highly specific and antigenic recombinant SCoV nucleocapsid (rSCoVN) protein, and to provide a basis for further researches on early diagnosis of SARS.
METHODS: Full length cDNA of SCoV nucleocapsid (SCoVN) protein was amplified through polymerase chain reaction (PCR) and cloned into yeast expression vector pPIC3.5K to construct plasmid of pPIC3.5K-SCoVN. The plasmid was linearized and then transformed into Pichia pastoris (P.pastoris) GS115 (His-Mut+) by electroporation. His+Mut+ recombinant strains were identified by PCR and cultivated on MM/MD plates. The influence of different factors on biomass and rSCoVN protein production during induction phase, such as various induction media, dissolved oxygen (DO) and different final concentrations of methanol, was subsequently studied. The expression level and activation were detected by SDS-PAGE and Western-blot respectively.
RESULTS: All of the recombinants were His+Mut+ after transformation of P.pastoris with linearized plasmids. The BMMY medium was optimal for recombinant ScoVN (rSCoVN) protein expression and growth of the recombinant strains. The final optimal concentration of methanol was 20 mL/L, the DO had a significant effect on rSCoVN protein expression and growth of recombinant strains. The rSCoVN protein expressed in recombinant strains was about 8% of the total cell protein, 520 mg/L of rSCoVN protein was achieved, and a maximum cell A at 600 nm of 62 was achieved in shake flask culture. The rSCoVN protein had a high specificity against mouse-anti-SARS-CoVN-mAb and SARS positive sera, but had no cross-reaction with normal human serum. The biological activity of rSCoVN expressed in P.pastoris was about 4-fold higher than that expressed in E.coli when the same rSCoVN protein quantity was used.
CONCLUSION: Active recombinant severe acute respiratory syndrome (SARS) coronavirus nucleocapsid (rSCoVN) protein can be successfully expressed in recombinant methylotrophic yeast P.pastoris GS115. The rSCoVN protein has a high specificity against SARS-CoVN-mAb and SARS positive sera, but has no cross-reaction with normal human serum. This provides a basis for further researches on the early diagnosis of SARS and the mechanism of SCoV.
-
Citation: Liu RS, Yang KY, Lin J, Lin YW, Zhang ZH, Zhang J, Xia NS. High-yield expression of recombinant SARS coronavirus nucleocapsid protein in methylotrophic yeast
Pichia pastoris . World J Gastroenterol 2004; 10(24): 3602-3607 - URL: https://www.wjgnet.com/1007-9327/full/v10/i24/3602.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i24.3602
The etiologic agent of SARS has been recently identified as a novel coronavirus (SARS-CoV, SCoV) causing respiratory and enteric diseases in humans and other animals[1-5]. The SCoV is a group of large, enveloped, positive single stranded RNA (ssRNA) viruses[6]. Sequence analysis has revealed that the phylogeny of SCoV has most of the characteristic features of a coronavirus, but it belongs to a new group different from all known coronaviruses[7]. The genome size is 29.725 kb in full-length, has 11 open reading frames (ORFs)[8]. The SCoV is the largest virus found in any of the RNA viruses, encoding 23 putative proteins, including four major structural proteins, nucleocapsid (N), spike (S), membrane (M), and small envelope (E). The S, M, E and N mature proteins all contribute to generating the host immune response as seen in transmissible gastroenteritis coronavirus[9], infectious bronchitis virus[10,11], pig respiratory coronavirus[12], and mouse hepatitis virus[13].
SARS is an infectious disease with a high potential for transmission due to close contacts. The outbreak of SARS over 25 countries around the world, such as China, Singapore, Canada, threatened people’s health throughout the world. Within a very short time, the disease became pandemic with new cases appearing in the rest of the world. However, it is not easy to differentiate SARS from other causes of pneumonia. Laboratory tests that can confirm a diagnosis of SCoV infection early in the course of the illness are therefore a critical clinical need[14]. Serology is a sensitive and specific diagnostic approach in the early stage of the disease. N protein plays an important role in reproduction and pathology reaction of SCoV. Moreover, the antigenicity of N protein is better than S protein[9,15,16]. To find an effective serologic diagnostic method, rSCoVN may be a perfect antigen.
Pichia pastoris (P.pastoris), a methylotrophic yeast, is an efficient host for recombinant protein production. The increasing popularity of this particular expression system can be attributed to several factors[17-19], such as the simplicity of techniques needed for the molecular genetic manipulation of P.pastoris, the ability of P.pastoris to produce foreign proteins at high levels, many eukaryotic posttranslational modifications, and the commercially available expression system.
Up to now, rSCoVN has been expressed in E.coli. However, the biological activity of the rSCoVN protein expressed in E.coli is not perfectly understood. High-yield intracellular expression in P.pastoris and activity analysis were investigated in this report. Western blot showed that the rSCoVN expressed in P.pastoris had a high specificity to mouse-anti-SCoVN-mAb and patient sera. The biological activity of rSCoVN expressed in P.pastoris was about 4-fold higher than that expressed in E.coli when the same rSCoVN protein quantity was used.
E.coli strains used for cloning the gene were TOP10F‘[proAB, lacIq, lacZ△M15, Tn10 (TetR)], and TG1 (supE hsd△ 5 thi△ (lac-proAB) F‘[traD36 PROAB+ lacIq, lacZ△M15] ). These two strains were cultivated in LB medium (5 g/L yeast extract, 10 g/L peptone, 5 g/L NaCl). P.pastoris GS115 (his-mut+) used as expression host was purchased from Invitrogen (California, USA). The following media were employed in cultivation of P.pichia cells under different conditions: YEPD (10 g/L yeast extract, 20 g/L peptone, 20 g/L dextrose), BMGY and BMMY (10 g/L yeast extract, 20 g/L peptone,100 mmol/L potassium phosphate, 13.4 g/L YNB, 4 × 10-5 biotin, 10 g/L glycerol , pH6.0), for BMMY, 5 mL/L filtersterilized methanol was added instead of 10 g/L glycerol. BMG and BMM (100 mmol/L potassium phosphate, 13.4 g/L YNB, 4 × 10-5 biotin, 10 g/L glycerol, pH6.0), for BMM, 5 mL/L filtersterilized methanol was added instead of 10 g/L glycerol. MGY and MM (13.4 g/L YNB, 4 × 10-5 biotin, 10 g/L glycerol), for MM, 5 mL/L filtersterilized methanol was added instead of 10 g/L glycerol. MD and MM agar plates (13.4 g/L YNB, 4 × 10-5 biotin, 20 g/L dextrose, 15 g/L agar), for MM, 5 mL/L filtersterilized methanol was added instead of 20 g/L dextrose. RDB and RDHB (20 g/L dextrose, 13.4 g/L YNB, 4 × 10-5 biotin, 0.05 g/L each of filtersterilized L-glutamic acid, L-lysine, L-leucine, L-isoleucine, L-methionine), for RDBH, 0.05 g/L filtersterilized histidine was added. RDB-G418 agar plates (RDB agar plates, 0.5-3 g/L G418 was added). T-vector was purchased from Takara (Dalian city, China), plasmid pPIC3.5K was purchased from Invitrogen (California, USA).
Construction of pPIC3.5K-SCoVN expression plasmid The standard recombinant DNA technologies[20] were used. The coding sequence of SCoVN protein was amplified from the T-SCoVN plasmid using the upstream primer (SCoVNFP: 5’-GGATCCACCATGTCTGATAATGGACCCC-3’) containing a BamH I site, in addition to a Kozak consensus sequence, and the reverse primer (SCoVNRP: 5’-GAATTCTTATGCCTGAGTT GAATCAG-3’) contained an EcoR I site immediately downstream of an inframe stop condon. The primers facilitated the subcloning of SCoVN protein coding sequence into pPIC3.5 K expression vector. The conditions for PCR were as follow: template was initially denatured at 94 °C, cultivated for 4 min, followed by 19 cycles, (each at 94 °C for 50 s, at 57 °C for 50 s, at 72 °C for 80 s), finally, a cycle was performed at 72 °C for 10 min. The PCR products were ligated to T-vector, then cut off by BamH I and EcoR I and the fragments were ligated into the same enzymes digested multiple cloning sites of pPIC3.5K expression vector. Finally, the clone was sequenced by 5’AOX primer and 3’AOX primer in the kit, and the fragments were confirmed to be inserted into the correct sites.
Transformation of P.pastoris and selection of his+Mut+ transformantsP.pastoris was transformed using electroporation protocol, competent cells were prepared as described[21]. Transformed DNA was linearized using Sac I leading to targeting of recombinant plasmids to the chromosomal his4 locus. Before transformation, the linearized DNA was desalted by gel extraction mini kit (Watson Biotechnologies, INC). Plasmid DNA (3-5 μg ) was mixed with 80 μL of competent cells and stored on ice for 5 min. Cells transferred to an ice-cold 0.2 cm electroporation cuvette. Transformation was performed using a BioRad GenePulser II. Parameters used were 7.5 KV/cm, 50 μF and 400 Ω. After pushed, 1.0 mL of ice-cold RDB liquid media was immediately added to the cuvette, and then incubated at 29 °C for 60 min in a shaking incubator (250-300 rm). Two aliquots of 400 μL each were plated on RDB-geneticin agar plates, the plates were incubated at 29 °C for 3-4 d. Using sterile toothpicks, the His+ and geneticin resistant transformants were picked in a regular pattern on MM and MD plates and incubated at 29 °C for 2-3 d. The His+Muts (methanol utilization slow) transformants were differentiated from his+Mut+ (methanol utilization plus) via comparison of patch growth rate on MM and MD plates.
PCR analysis of P.pastoris recombinant transformants The primers used were as follows. 5’AOX1: 5’-GACTGGTTCCAAT TGACAAGC-3’;3’AOX1:5’-GCAAATGGCATTCTGACATCC-3’. The genomic DNA was isolated as described in multi-mopy Pichia expression kit. PCR amplification was performed as follows: initial denaturation at 95 °C for 4 min, followed by 34 cycles, (each at 94 °C for 60 s, at 55 °C for 60 s, at 72 °C for 150 s) and a final extension at 72 °C for 10 min. The genomic DNAs isolated from recombinant P.pastoris transformed with parent plasmid and P.pastoris GS115 were used as control for PCR[21].
Shake-flask cultivation of P.pastoris and intracellular expression of rSCoVN The recombinant P.pastoris strains judged by PCR analysis were grown in 15 mL BMGY medium at 29 °C, until the final cell OD at 600 nm reached 2-6 (log-phase growth). The cells were harvested by centrifuging at 1500 r/min for 6 min at room temperature, cell pellets were resuspended to a cell A at 600 nm of 1.5 in BMMY medium and induced at 29 °C in a shaking incubator (250-300 rm). A 100 mL/L of methanol was added to a final concentration 5 mL/L every 24 h to maintain induction. Induced P.pastoris transformed with the parent vector was used as a control for background intracellular expression. At each time (about 12-24 h), samples were withdrawn to analyse expression level, activity and to detect A at 600 nm.
Optimization of culture and induction protocol in shake flasks The highly expressed recombinant strains judged by BMGY/BMMY media firstly were grown in different media, and then induced in relevant media. At last, the A and the final methanol concentration were optimized during induction phase. At each time during induction, samples were withdrawn to analyse expression level and the biomass of the recombinant strains.
Lysis of cells and detection of proteins Yeast cells induced to express rSCoVN protein were harvested, the cells were washed with sterile water, and then harvested and stored at -80 °C. Frozen yeast cells were removed from storage and thawed for approximately 3 h at room temperature, breaking buffer (25 mmol/L sodium phosphate, 150 mmol/L sodium chloride, 1 mmol/L EDTA, 1 mmol/L DTT) was added, the cells (6 mL breaking buffer/g wet cells) were resuspended and then stirred for 15 min, followed by approximately 18 h at 4 °C, then disrupted by four passes through a sanitized APV Gaulin 30 CD homogenizer at chamber pressures of 12000 to 14000 psi, resulting in 95% cell disruption. About 10 μL breaking sample mixed with 10 μL ddH20 and 10 μL SDS-PAGE sample buffer[22] were boiled for 5-10 min, then centrifuged at 12000 g for 10 min at room temperature, and 10 μL of the supernate was applied to SDS-PAGE, and separated on 100 g/L polyacrylamide gels with 50 g/L stacking gel. For protein estimations, Coomassie-stained SDS-gels were analyzed by densitometry using UVI.
Western blot analysis of rSCoVN protein After equilibration of gels and nitrocellulose membranes in transfer buffer (25 mmol/L Tris·HCl, 192 mmol/L glycine, 3.5 mmol/L SDS, 200 mL/L methanol), the protein was electroblotted onto a membrane (Amersham Pharmacia Biotech Hoefer TE 70 Series Semiphor Semi-Dry transfer Units, 0.8 mA/cm2 30 min)[23]. After the membrane (50 g/L fat free milk/TN buffer (10 mmol/L Tris·HCl, 150 mmol/L NaCl, pH8.0) was blocked, it was probed with a mouse-anti-SCoVN-mAb (dilution 1:1000, 50 g/L fat free milk/TN buffer or human serum (dilution 1:100 in 5% fat free milk/TN buffer; 20-25 °C, 1 h), followed by a sheep-anti-mouse or sheep-anti-human IgG coupled to alkaline phosphatase (dilution: 1:10000 in 50 g/L fat-free milk/TN buffer; 20-25 °C, 1 h). Bound antibodies were detected using 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) and nitro-blue tetrazolium (NBT) in 100 mmol/L NaCl, 5 mmol/L MgCl2, 100 mmol/L Tris. HCl buffer, pH9.5[7] as substrates.
Quantitative activity analysis of rSCoVN proteins expressed in P.pastoris and E.coli The biological activity of rSCoVN proteins expressed in P.pastoris and E.coli was compared using Western blot. The purified protein expressed in P.pastoris and E.coli was diluted by 4-fold gradient, and 12, 3, 0.75, 0.18 µg proteins were added to each well of SDS-PAGE respectively, then probed with a mouse-anti-SCoVN-mAb.
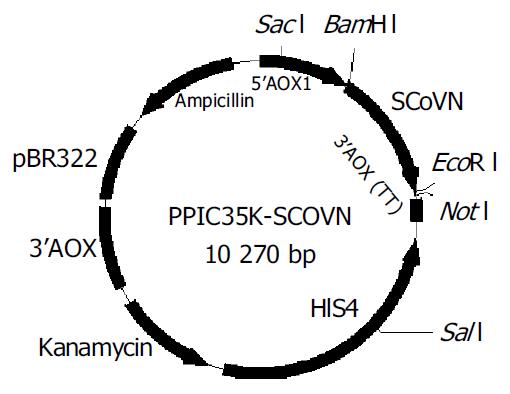
The coding sequences for SCoVN gene were amplified from T-SCoVN plasmid by PCR using primers (SCoVNFP and SCoVNRP) incorporating 5’BamH I and 3’EcoR I, and subcloned into the BamH I and EcoR I sites of the pPIC3.5K expression plasmid (Figure 1). Then the constructed expression plasmid was sequenced and proved to be correct. The SCoVN protein coding sequence was under the control of the AOX1 promoter.
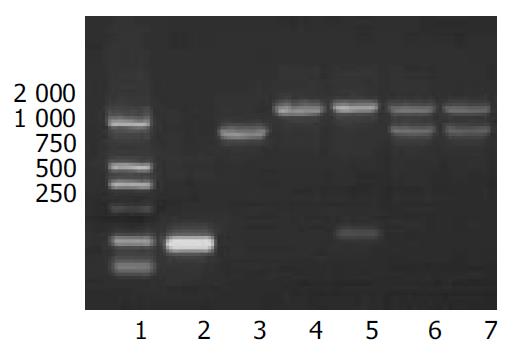
pPIC3.5K-SCoVN or pPIC3.5K was digested with Sac I to linearize the plasmids, and then electroporated into P.pastoris GS115 (his+mut+) respectively. Twenty geneticin-resistant colonies were cultivated on MD and MM plates. They were all His+Mut+, because they all had the same growth rate. The geneticin-resistant colonies were grown in YPED for 24 h, then the genomic DNA was purified, PCR amplification of SCoVN gene was carried out with 5’AOX1 primer and 3’AOX1 primer. Twenty transformants each had the alcohol oxidase gene and SCoVN gene. The result demonstrated that all geneticin-resistant colonies were His+Mut+, identical to the result identified on MD and MM plates. The SCoVN protein coding sequence was correctly integrated into the P.pastoris genome in the positive recombinants via a single crossover (Figure 2: lanes 5 and 6).
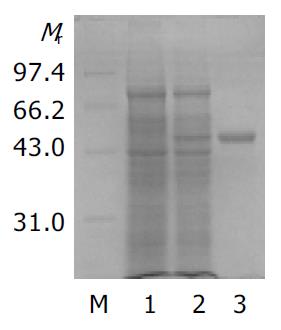
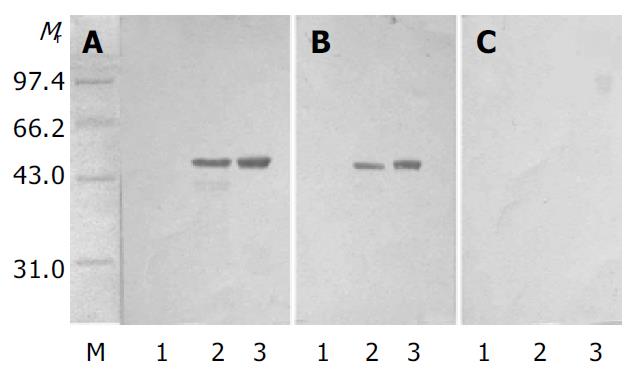
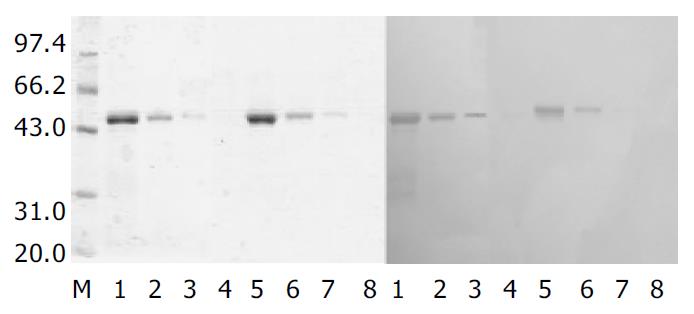
A single recombinant colony (His+Mut+) was used for expression study in shake flask expression[21]. After one day of cell growth on BMGY medium at 30 °C, 250 rm, then the BMGY medium was changed to BMMY medium. The cells were fed with 100 mL/L methanol to 5 mL/L (final concentration) every day. The strains transformed by parent plasmid pPIC3.5K were taken as control. Samples were withdrawn after 60 h. SDS-PAGE showed that heterologous rSCoVN protein was successfully expressed in P.pastoris, the rSCoVN protein expression level was about 6% of the total cell protein (Figure 3), a 45 ku protein band could be seen in the positive recombinant and there was no 45 ku protein band in control. To confirm the 45 ku protein was the SCoVN protein, 45 ku protein was detected by Western blot with mouse-anti-SCoVN protein mAb, SARS positive serum and negative serum. The results showed that a single positive reaction band at about 45 ku (Figure 4) could be seen in the nitrocellulose membrane. The band of the same molecular mass could not be detected in the induced recombinant strain transformed pPIC3.5K vector by Western blot analysis. The results showed that the recombinant protein was successfully expressed and had a high-specificity and good-antigenicity against SCoVN-Ab and SARS positive serum, but had no reaction with normal human serum.
The biologic activities of rSCoVN protein expressed in P.pastoris and E.coli were compared using Western-blot. As shown in Figure 5, both rSCoV proteins were able to react with mouse-anti-SARSN-mAb. However, the rSCoVN protein expressed in P.pastoris appeared to have a more potent activity than that in E.coli. Dose-response studies demonstrated that the biological activity of rSCoVN expressed in P.pastoris was approximately 4-fold higher than that in E.coli when the same rSCoVN protein quantity was used. But the Western blot and SDS-PAGE demonstrated that there was no glycosylation in the rSCoVN protein expressed in P.pastoris, because the rSCoVN protein expressed in P.pastoris had the same molecular weight as in E.coli. So rSCoVN protein expressed in P.pastoris had more application potential as a diagnotic agent.
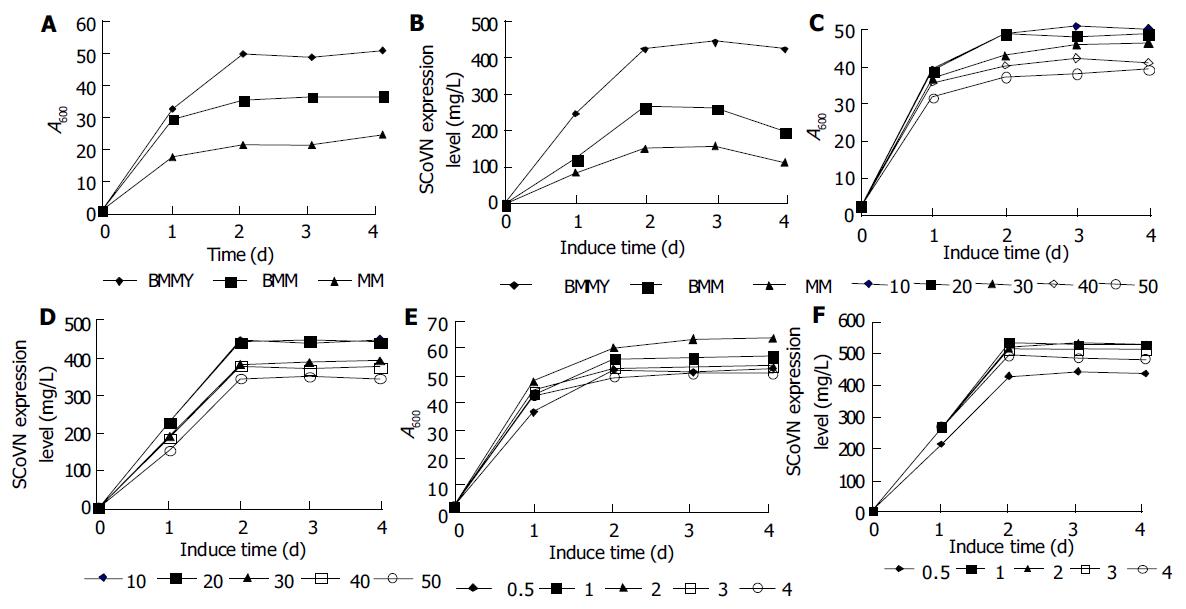
Comparison of the effect of various media on yeast growth and rSCoVN expression To investigate the effect of different media on rSCoVN expression level and the growth of recombinant strains, we compared the rScoVN expression level and biomass in different induction media such as BMMY, BMM and MM media in 20 mL scale. rSCoVN protein expression in P.pastoris was performed with a two-phase culture method: biomass production using glycerol as a solely carbon source and specific induction by methanol respectively in BMMY, BMM and MM media. Figure 6A, B show the biomass and rSCoVN protein expression level in each medium at different induction stages. The recombinant strains reached the stationary phase after 2 d of induction, and rSCoVN protein expression level also reached the maximum. A 2-fold difference in biomass and 3.5-fold difference in expression level were noticeable between BMMY and MM media. Difference in biomass and expression was also found between BMMY and BMM media. BMMY medium was optimal for induction expression.
The effect of DO in media on growth and rSCoVN protein expression of P.pastoris was also investigated. Different volume media (10, 20, 30, 40 and 50 mL) were loaded in 100 mL flasks. The different volume of the media loaded in flasks meant different DO. The more the medium was loaded, the less the DO was, and it should be noted that the shape was identical. Figure 6C, D show that the DO had an obvious effect on the yeast growth and expression of rSCoVN. The rSCoVN protein expression level was the same in the flask loaded 10 mL or 20 mL medium, because the DO was efficient for growth and rSCoVN protein expression of the recombinant strains in the flask, so the growth and rSCoVN expression level of the recombinant protein were very high, over 20% higher than others. When the volume of medium was increased(above 20 mL), the DO decreased, and the growth and expression level were decreased. So the DO was an important factor for the growth and rSCoV protein expression of the recombinants.
To find an optimum methanol feeding protocol, different volumes of 100% methanol were added to the media twice a day instead of once a day to decrease the resulting concentration shifts in the BMMY medium. Figure 6E, F show that when the methanol feeding was below 20 mL/L, and the concentration of methanol was increased, the biomass was also increased, while the rSCoVN protein expression level was the same at the methanol feeding of 10 mL/L and 20 mL/L, but was increased about 25% than that at the methanol feeding of 5 mL/L. Above the methanol feeding of 20 mL/L, the higher the methanol was fed, the lower the biomass and expression level were. So feeding with 20 mL/L methanol twice a day was considered an optimal feeding strategy.
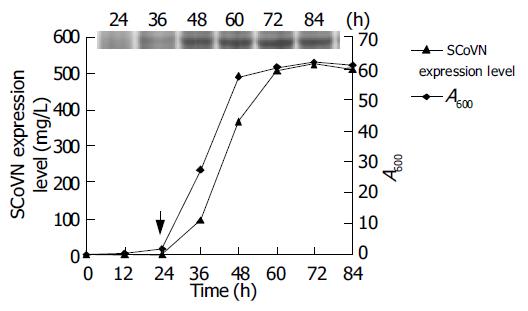
After the optimal conditions for yeast growth and rSCoVN protein expression were found, the kinetics of the growth and expression were examined. The results are shown in Figure 7. The rSCoVN protein could be detected readily by SDS-PAGE after 12 h of induction (the inserted panel), rSCoVN protein expression was evident when cells entered the logarithmic growth state and peaked when cell growth reached a steady state and became more or less constant thereafter. The magnitude of expression level reached 526 mg/L. The rSCoVN protein expression was about 8% of the total cell protein, and the maximum cell OD at 600 nm of 62 was achieved. So the optimal harvesting time of cells was 48-60 h after induction. The insert in the panel showed the accumulation of rSCoVN protein by SDS-PAGE during the course of the 84 h experiment.
The P.pastoris expression system has gained acceptance as an important host organism for the production of foreign proteins as illustrated by the fact that a number of proteins synthesized in P.pastoris are tested for use as pharmaceuticals in clinic. IGF-1 as a treatment for amyotrophic lateral sclerosis and human serum albumin (HSA) in a serum replacement product have passed clinical trials. Another protein, hepatitis B surface antigen, has been currently available on the market as a subunit vaccine against hepatitis B virus[24,25].
The etiologic agent of severe acute respiratory syndrome (SARS), has been recently identified as a novel coronavirus causing respiratory and enteric diseases in humans and other animals[1-5]. SARS is an infectious disease with a high potential for transmission due to close contacts. So it is critical to find a perfect diagnostic agent. N protein plays an important role in reproduction and pathologic reaction of SCoV, and its antigenicity is better than spike protein. So N protein may be the perfect antigen.
In the present study, active rSCoVN was highly expressed in P.pastoris. Compared with various media, the BMMY medium was optimal for rSCoVN expression. A 2-fold difference in biomass and 3.5-fold difference in expression level were noticeable between BMMY and MM media. Difference in biomass and expression was also presented between BMMY and BMM media. The result also revealed that peptone, yeast extracts and steady pH had prominent effects on the growth and expression of recombinant P.pastoris. The buffered medium could maintain a stable pH value, and benefit the absorption and use of the nutrients. Moreover, yeast extracts, peptone are rich in peptides, amino acids, vitamins and trace elements. These compounds could enhance the biomass and energy for foreign protein synthesis[25].
The effect of DO on the growth and expression of rSCoVN revealed that DO was an important factor for yeast growth and expression of recombinant protein. While microorganisms growing on carbohydrates use molecular oxygen mainly for respiration. Oxygen availability could influence the production of proteins, both in prokaryotic cells[26] and in yeast[27]. Yeasts growing on methanol might also require a substantial amount of oxygen for the initial oxidation of methanol to formaldehyde[28]. All methanol taken up by the cells is oxidized to formaldehyde in a coupled reaction involving alcohol oxidase (AOX) and catalase (CAT) in peroxisomes. These reactions use molecular oxygen as an ultimate electron acceptor. Foreign protein synthesis needs abundant energy, so it is important to maintain a relatively high DO for expressing foreign protein in Pichia pastoris.
For the expression of protein using AOX1 promoter, it is important to keep the methanol level within a relatively narrow range. In bioreactors it could be achieved by different methods[29]. In shake flasks, a technique for online monitoring of methanol concentration was introduced[30]. The ordinary methanol feeding protocol without any complicated devices, still has not been reported. So we established an empirically feeding protocol for our recombinant strains, which worked without any measuring devices. A methanol feeding of 20 mL/L, final concentration) was optimal for yeast growth and expression of SCoVN protein. Slightly reduced growth and expression while methanol feeding below or above 20 mL/L was probably due to the limited carbon or the toxic effect of accumulated methanol[30].
Western blot demonstrated that rSCoVN protein had a high-specificity and good-antigenicity against SCoVN-Ab and SARS positive serum, and no reaction with normal human serum. The biological activity of rSCoVN expressed in P.pastoris was about 4-fold higher than that expressed in E.coli when the same rSCoVN protein quantity was used. The preliminary results indicate that the conformation of rSCoVN protein expressed in P.pastoris is almost the same as the natural SCoVN protein, and the rSCoVN protein produced may be suitable for the detection of anti-SCoV in diagnostic assay.
| 1. | Poutanen SM, Low DE, Henry B, Finkelstein S, Rose D, Green K, Tellier R, Draker R, Adachi D, Ayers M. Identification of severe acute respiratory syndrome in Canada. N Engl J Med. 2003;348:1995-2005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 771] [Cited by in RCA: 788] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 2. | Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt GM, Ahuja A, Yung MY, Leung CB, To KF. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986-1994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1563] [Cited by in RCA: 1609] [Article Influence: 70.0] [Reference Citation Analysis (1)] |
| 3. | Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319-1325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2097] [Cited by in RCA: 2170] [Article Influence: 94.3] [Reference Citation Analysis (0)] |
| 4. | Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953-1966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3147] [Cited by in RCA: 3095] [Article Influence: 134.6] [Reference Citation Analysis (0)] |
| 5. | Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967-1976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3277] [Cited by in RCA: 3345] [Article Influence: 145.4] [Reference Citation Analysis (0)] |
| 6. | Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763-1767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1209] [Cited by in RCA: 1366] [Article Influence: 59.4] [Reference Citation Analysis (0)] |
| 7. | Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, Khattra J, Asano JK, Barber SA, Chan SY. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399-1404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1538] [Cited by in RCA: 1564] [Article Influence: 68.0] [Reference Citation Analysis (5)] |
| 8. | Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Peñaranda S, Bankamp B, Maher K, Chen MH. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394-1399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1864] [Cited by in RCA: 1895] [Article Influence: 82.4] [Reference Citation Analysis (4)] |
| 9. | Gómez N, Carrillo C, Salinas J, Parra F, Borca MV, Escribano JM. Expression of immunogenic glycoprotein S polypeptides from transmissible gastroenteritis coronavirus in transgenic plants. Virology. 1998;249:352-358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 68] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 10. | Jackwood MW, Hilt DA. Production and immunogenicity of multiple antigenic peptide (MAP) constructs derived from the S1 glycoprotein of infectious bronchitis virus (IBV). Adv Exp Med Biol. 1995;380:213-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 11. | Ndifuna A, Waters AK, Zhou M, Collisson EW. Recombinant nucleocapsid protein is potentially an inexpensive, effective serodiagnostic reagent for infectious bronchitis virus. J Virol Methods. 1998;70:37-44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 12. | Callebaut P, Enjuanes L, Pensaert M. An adenovirus recombinant expressing the spike glycoprotein of porcine respiratory coronavirus is immunogenic in swine. J Gen Virol. 1996;77:309-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 13. | Homberger FR. Nucleotide sequence comparison of the membrane protein genes of three enterotropic strains of mouse hepatitis virus. Virus Res. 1994;31:49-56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 14. | Riley S, Fraser C, Donnelly CA, Ghani AC, Abu-Raddad LJ, Hedley AJ, Leung GM, Ho LM, Lam TH, Thach TQ. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961-1966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 807] [Cited by in RCA: 663] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 16. | Daginakatte GC, Chard-Bergstrom C, Andrews GA, Kapil S. Production, characterization, and uses of monoclonal antibodies against recombinant nucleoprotein of elk coronavirus. Clin Diagn Lab Immunol. 1999;6:341-344. [PubMed] |
| 17. | Sreekrishna K, Brankamp RG, Kropp KE, Blankenship DT, Tsay JT, Smith PL, Wierschke JD, Subramaniam A, Birkenberger LA. Strategies for optimal synthesis and secretion of heterologous proteins in the methylotrophic yeast Pichia pastoris. Gene. 1997;190:55-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 290] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 18. | Hasslacher M, Schall M, Hayn M, Bona R, Rumbold K, Lückl J, Griengl H, Kohlwein SD, Schwab H. High-level intracellular expression of hydroxynitrile lyase from the tropical rubber tree Hevea brasiliensis in microbial hosts. Protein Expr Purif. 1997;11:61-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 91] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 19. | Hollenberg CP, Gellissen G. Production of recombinant proteins by methylotrophic yeasts. Curr Opin Biotechnol. 1997;8:554-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 129] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 20. | Sambrook J. Molecular Cloning: A Laboratory manual, 2nd ed. cold spring harbor: cold spring harbor laboratory press, 1989. . |
| 21. | Invitrogen, Instruction manual for multi-copy pichia expres-sion kit-version F, 1999-2002. . |
| 22. | Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185380] [Cited by in RCA: 189403] [Article Influence: 3382.2] [Reference Citation Analysis (1)] |
| 23. | Amersham pharmacia biotech. Hoef TE 70 Series semiphor semi-Dry Transfer Units User Manual,1998. . |
| 24. | Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24:45-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1396] [Cited by in RCA: 1378] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 25. | Cregg JM, Vedvick TS, Raschke WC. Recent advances in the expression of foreign genes in Pichia pastoris. Biotechnology (N Y). 1993;11:905-910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 446] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 26. | Shioya S, Morikawa M, Kajihara Y, Shimizu H. Optimization of agitation and aeration conditions for maximum virginiamycin production. Appl Microbiol Biotechnol. 1999;51:164-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 27. | Hallborn J, Gorwa MF, Meinander N, Penttilä M, Keränen S, Hahn-Hägerdal B. The influence of cosubstrate and aeration on xylitol formation by recombinant Saccharomyces cerevisiae expressing the XYL1 gene. Appl Microbiol Biotechnol. 1994;42:326-333. [PubMed] |
| 28. | Veenhuis M, Van Dijken JP, Harder W. The significance of peroxisomes in the metabolism of one-carbon compounds in yeasts. Adv Microb Physiol. 1983;24:1-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 148] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 29. | Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24:45-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1396] [Cited by in RCA: 1378] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 30. | Boettner M, Prinz B, Holz C, Stahl U, Lang C. High-throughput screening for expression of heterologous proteins in the yeast Pichia pastoris. J Biotechnol. 2002;99:51-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 86] [Article Influence: 3.6] [Reference Citation Analysis (1)] |
Edited by Wang XL and Zhu LH Proofread by Xu FM