Published online Dec 1, 2004. doi: 10.3748/wjg.v10.i23.3433
Revised: April 29, 2003
Accepted: May 9, 2003
Published online: December 1, 2004
AIM: To establish the DNA methylation patterns of the promoter CpG islands of 14 “drug-resistance” genes in hepatocellular carcinoma (HCC).
METHODS: The methylation specific polymerase chain reaction in conjunction with sequencing verification was used to establish the methylation patterns of the 14 genes in the liver tissues of four healthy liver donors, as well as tumor and the paired non-cancerous tissues of 30 HCC patients.
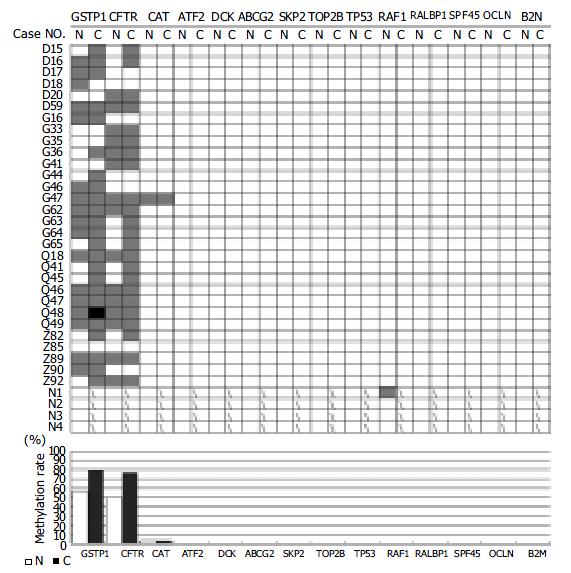
RESULTS: While 11 genes (ATP-binding cassette, sub-family G (WHITE), member 2(ABCG2), activating transcription factor (ATF2), beta-2-microglobulin (B2M), deoxycytidine kinase (DCK), occludin (OCLN), v-raf-1 murine leukemia viral oncogene homolog (RAF1), ralA binding protein 1 (RALBP1), splicing factor (45 kD) (SPF45), S-phase kinase-associated protein 2 (p45) (SKP2), tumor protein p53 (Li-Fraumeni syndrome) (TP53) and topoisomerase (DNA) II beta (TOP2B)) maintained the unmethylated patterns, three genes displayed to various extents the hypermethylation state in tumor tissues in comparison with the normal counterparts. The catalase (CAT) was hypermethylated in tumor and the neighboring non-cancerous tissue of one case (3.3%). Both glutathione S-transferase pi (GSTpi) (80%, 24/30 in tumor and 56.7%, 17/30 in the paired non-cancerous tissues) and cystic fibrosis transmembrane conductance regulator, ATP-binding cassette (sub-family C, member 7) (CFTR) (77%, 23/30 in tumor and 50%, 15/30 in the paired non-cancerous tissues) genes were prevalently hypermethylated in HCC as well as their neighboring non-cancerous tissues. No significant difference in the hypermethylation occurrence was observed between the HCC and its neighboring non-cancerous tissues.
CONCLUSION: Hypermethylation of promoter CpG islands of both CFTR and GSTpi genes occurs prevalently in HCC, which may correlate with the low expression of these two genes at the mRNA level and has the profound etiological and clinical implications. It is likely to be specific to the early phase of HCC carcinogenesis.
- Citation: Ding S, Gong BD, Yu J, Gu J, Zhang HY, Shang ZB, Fei Q, Wang P, Zhu JD. Methylation profile of the promoter CpG islands of 14 “drug-resistance” genes in hepatocellular carcinoma. World J Gastroenterol 2004; 10(23): 3433-3440
- URL: https://www.wjgnet.com/1007-9327/full/v10/i23/3433.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i23.3433
Hepatocellular carcinoma (HCC) is one of the most threatening malignancies, occurring prevalently in China[1]. Its poor prognostic prospects have not only been attributed to the rapid advancing nature as well as the notorious metastasizing potential of the primary lesions, but also resulted from its refractoriness to the conventional chemotherapeutic practices[2-4]. Altered expression of the crucial genes, even prior to the relevant treatment, is common in cancer cells, including HCC. The intrinsic drug resistance, reflected by the changes in the expression profile of the key genes in relevant pathways, including apoptosis, cell cycle progression, DNA repairs, etc., is common in cancer cells, which certainly contributes to the more rapid growth ability of cancerous cells than the normal counterparts, in the adverse environments in particular. Furthermore, cancer cells can also acquire resistance to chemotherapeutic remedies during treatment. It has been regarded as the key mechanism for the post-remission return of the full-blew cancer of same origin. Therefore, a long-standing challenge in cancer field is the underlying mechanisms of the drug resistance nature of tumors. The genetic defects leading to the intrinsic drug resistance have been well-established, but the mechanisms at the epigenetic levels, DNA methylation in particular resulting in the changes in expression, have only gained the justified recognition recently[5].
Over 50% of the protein coding genes are marked with the CpG rich segment at their 5’ end, the methylation status of which profoundly affects the transcription status of the genes[6]. Biochemically, the methylated CpG affects the sequence-specific DNA-protein interactions by eliminating the otherwise binding of the transcription factors to their cognate cis-elements, while unfolding the cascade of reactions leading to chromatin condensation initiated by the binding to the methylated CpG by members of the methyl CpG binding protein family (MBD). Hypermethylation of the promoter CpGs has been linked to the long-term transcription-silencing status of DNA segments, including genes as well as the transposon-like-repetitive sequences in cells[7,8]. The genome-wide hypomethylation has been regarded contributive to activation of transcription of the otherwise silenced transposon like repetitive sequences (such as the Alu and LINE repeats in mammals) and instrumental to the transposition mediated loss of the genome stability during cell transformation[9-11]. The CpG islands of several tumor suppressor genes, which are hypomethylated and express in normal cells, became frequently hypermethylated in almost all the cancers investigated, contributing to the loss of function of the tumor suppressor genes in cancer. On the other hand, the reverse processes, such as hypomethylation of CpG islands may also result in transcription activation of the otherwise inert genes[12].
In our previous studies on HCC[13], we have assessed a number of genes in the category of “drug-resistance” genes, including the O-6-methylguanine-DNA methyltransferase (MGMT) gene encoding an enzyme responsible for the cell’s resistance to the alkylating type of chemotherapeutic drugs, and mutL homolog 1, colon cancer, nonpolyposis type 2 (hMLH1), a mismatch repairing gene[14]. The former is frequently methylated while the later maintains the unmethylated status in HCC tissues in comparison with the normal liver tissues. In this study, we extended our profiling efforts in the same group of HCC patients to other 14 “drug-resistance” genes, 13 of which were touched for the first time. This specifically oriented survey to “drug-resistance” should provide valuable insights into our understanding of the underlying mechanisms for the development of intrinsic resistance of HCC to chemotherapeutic drugs, and have important etiological implications to carcinogenesis of HCC.
With the informed consent of all patients and approval of the ethics committee, the samples of tumors were collected from HCC patients (n = 30) during operation. The pathological classification of tumor tissues was carried out, and the stage of each case was graded according to the WHO classification[15]. Total genomic DNA was extracted from frozen tissue specimens (50-100 mg) according to a standard protocol with some modifications[16,17]. Frozen pulverized powders of the specimens were re-suspended with 2 mL lysis buffer containing 50 mmol/L Tris-HCl pH 8.0, 50 mmol/L EDTA, 10 g/L SDS, 10 mmol/L NaCl plus 100 μg/mL boiling-treated RNase A (Sigma, USA). Following 1-h incubation at 37 °C, proteinase K (Roche, USA) was added to the cellular lysates for a final concentration of 100 μg/mL, and the digestion was carried out at 55 °C for 2 h. Organic extractions with a half volume of phenol/chloroform/isoamyl alcohol (1:1:0.04) were repeatedly carried out until no visible interphase remained after centrifugation. DNA was precipitated from the aqueous phase in the presence of 0.3 mol/L NaOAc (pH 7.0) and two and a half volumes of ethanol, washed once in 700 mL/L ethanol, dissolved at 65 °C for 30 min with 0.2-0.4 mL TE (10 mmol/L Tris-HCl pH 7.4 and 1 mmol/L EDTA) and stored at 4 °C until use. The DNA concentrations were calculated according to the A260 nm readings.
We used “drug resistance” as the key word to search from the NCBI LocusLink database (http://www.ncbi.nlm.nih.gov/ LocusLink/list.cgi). The sequence of the coding region (63 targets) plus 5 kb segments at each ends were downloaded, and subjected to search for the existence of the CpG island. The parameters were set as OBS/EXP (the minimum average observed to expected ratio of C plus G to CpG) = 0.6, MINPC (the minimum average percentage of G plus C) = 50, and LENGTH (the minimum length that a CpG island has to be) = 200 bp (http://www.ebi.ac.uk/emboss/cpgplot/). Thirty six genes were found containing the promoter CpG island (Table 1). Among this list 14 genes were selected for the methylation profiling, as they represent in different categories for the functions and properties (Table 2). The primer pairs (Table 3) for methylation specific polymerase chain reaction (MSP) in this report were designed according to the same principle with assistance of the web server for identification of the CpG islands (http://http://www.ebi.ac.uk/emboss/cpgplot/index.html) and the primer design software (http://micro-gen.ouhsc.edu/cgi-bin/ primer3_http://www.cgi) (Table 3).
| Genes | Location relative to transcription start site | Genes | Location relative to transcription start site |
| ABCC4 | -487 to +466 | GSTP1 | -391 to +91 |
| ABCC1 | -356 to +406 | hMLH1 | -72 to +393 |
| ABCC13 | +138 to +386 | KIAA1337 | -553 to +413 |
| ABCC3 | -96 to +358 | MAPK14 | -371 to +319 |
| ABCB11 | +1 255 to +1 565 | OCLN | -9 to +967 |
| ABCC5 | -219 to +87 | POLB | -209 to +34 |
| ABCC6 | -229 to +89 | RAF1 | -466 to +399 |
| ABCC8 | -185 to +531 | RALBP1 | -901 to +325 |
| ABCG2 | -741 to -17 | RRM2 | -718 to +426 |
| ATF2 | -418 to +545 | SKP2 | -663 to +182 |
| B2M | -170 to +83 | SLC19A1 | -719 to +268 |
| BCAR3 | -328 to +126 | SLC22A1L | -265 to +158 |
| BCL2 | -7 to +573 | SPF45 | -547 to +92 |
| BIRC4 | -527 to -235 | SRI | -251 to +257 |
| CAT | +35 to +395 | SSA2 | -195 to +152 |
| CFTR | -515 to -55 | TOP2A | -123 to +506 |
| DCK | -267 to +155 | TOP2B | -805 to +393 |
| ERBB2 | +173 to +534 | TP53 | +130 to +337 |
| Resistance mechanism | Genes and ref. | Function |
| Plasma membrane | ABCG2 | Belongs to the abc transporter family, appears to play a major role in the multidrug resistance |
| transporters/pump | phenotype of MCF-7 breast cancer cell line. When over-expressed, the transfected cells become | |
| decrease or increase to | resistant to mitoxantrone, daunorubicin and doxorubicin, display diminished intracellular | |
| concentration | accumulation reduce intracellular drug of daunorubicin, and manifest an | |
| ATP-dependent increase in the efflux of rhodamine | ||
| CFTR | Belongs to the abc transporter family. MRP subfamily, involved in the transport of chloride ions | |
| RALBP1 | Activates specifically hydrolysis of GTP bound to RAC1 and CDC42. Mediates ATP-dependent | |
| transport of S-(2,4-dinitrophenyl)-glutathione (DNP-SG) and doxorubicin(DOX) and is the major | ||
| ATP-dependent transporter of glutathioneconjugates of electrophiles (GS-E) and DOX in | ||
| erythrocytes. Can catalyze transport of glutathione conjugates and xenobiotics, and may | ||
| contribute to the multidrug resistance phenomenon | ||
| Transcription factor | ATF2 | Belongs to the bZIP family, ATF subfamily, binding the cAMP response element (CRE) |
| and/or tumor suppressor | (consensus sequence: 5’GTGACGT(A/C)(A/G)-3') as a transcription activator | |
| TP53 | Acts as a tumor suppressor in many tumor types, involved in cell cycle regulation | |
| as a transcription activator that acts to negatively regulate cell division by controlling | ||
| a set of genes required for this process | ||
| The enzymes involved in | CAT | Belonging to the catalase family ,serves to protect cells from the toxic effects of |
| drug activation or inactivation | hydrogen peroxide | |
| metabolism abnormality | GSTP1 | Belongs to the GST superfamily. Pi family; Conjugation of reduced glutathione to |
| a wide number of exogenous and endogenous hydrophobic electrophiles | ||
| TOP2B | Control of topological states of DNA by ATP-dependent breakage, passage and | |
| rejoining of double-stranded DNA | ||
| DCK | Required for the phosphorylation of several deoxyribonucleosides and certain | |
| nucleoside analogs widely employed as antiviral and chemotherapeutic agents | ||
| Cell signaling alteration and | RAF1 | Belongs to the Ser/Thr family of protein kinases. RAF subfamily. Involved in |
| cell cycle checkpoints alteration | the transduction of mitogenic signals from the cell membrane to the nucleus as a | |
| part of the Ras-dependent signaling pathway from receptors to the nucleus | ||
| SKP2 | Involved in regulation of G1/S transition as substrate recognition component of the | |
| SCF (SKP1-CUL1-F- box protein) E3 ubiquitin ligase complex which mediates the | ||
| ubiquitination and subsequent proteasomal degradation of target proteins involved in | ||
| cell cycle progression, signal transduction and transcription | ||
| Unclear mechanisms | B2M | Beta-2-microglobulin is the beta-chain of major histocompatibility complex class I |
| molecules, Containing 1 immunoglobulin-like domain | ||
| OCLN | May play a role in the formation and regulation of the tight junction (TJ) | |
| paracellular permeability barrier | ||
| SPF45 | Involved in nuclear mRNA splicing, via spliceosome |
| GenBank No. | Gene symbol | Sense 5’-3’ | Antisense 5’-3’ | Position | Size(bp) | |
| NT_016354 | ABCG2 | m | TGTCGCGTTGAGTCGTTA | AACGTCCCCGATACTTCG | -698 to -464 | 235 |
| U | TGTGTTTTGTTGTGTTGAGTTGT | TCACTCTAATTCATTCCATTCAATC | -591 to -457 | 135 | ||
| NT_005403 | ATF2 | m | GGGTCGGAATAACGAACG | ATCACCTCGAATACTCCTAACG | 230 to 375 | 146 |
| U | GGAGGGGTTGGAATAATGAAT | CCCATTTCCCATCACCTCAA | 220 to 379 | 160 | ||
| NT_010194 | B2M | m | ATTTGGTATTGCGTCGTTG | ACGAAACGAAACATCTCGAC | -128 to 23 | 151 |
| U | TTTTTAATTTGGTATTGTGTTGTTG | AACTCACACTAAATAACCTCCAAAC | -134 to 83 | 217 | ||
| NT_009237 | CAT | m | AGTAGCGGGTCGCGTAG | AACCACCCGAACCTATCG | 118 to 372 | 255 |
| U | GGAAGGAGTAGTGGGTTGTGT | AACCACCCAAACCTATCACA | 112 to 372 | 261 | ||
| NT_007933 | CFTR | m | AGAGGTCGCGATTGTCGTT | CGACTTTCTCCACCCACTACG | -316 to -114 | 203 |
| U | TTAAAGAGAGGTTGTGATTGTTGTT | TCCTTCACTCCCTCACCA | -322 to -174 | 149 | ||
| NT_006216 | DCK | m | TATACGCGCGGTTTCGT | CGCCGACGAATATCGAA | -168 to 27 | 195 |
| U | TTTTTTGTTATATGTGTGGTTTTGT | TACCCCTCAACCCTCACC | -176 to -75 | 102 | ||
| NT_078018 | OCLN | m | TGCGTTCGTTAGGTGAGC | CGAATCCCAACTCGAAAACG | 538 to 753 | 216 |
| U | GTTAGGTGTGTTTGTTAGGTGAGT | CACACCTCTCTAATTCCCACA | 532 to 772 | 241 | ||
| NT_022517 | RAF1 | m | TCGGTCGTTTTGGAAGTC | CCCTAAAACGCGAAACG | -72 to 180 | 252 |
| U | GGTTTGGTTGTTTTGGAAGTT | CACCAAATATAACCACCTCCCACT | -2 to 183 | 185 | ||
| NT_010859 | RALBP1 | m | GGGTAAGTCGTTCGTTTTCG | CCTCTCCGCTCAAACGACT | -495 to -215 | 281 |
| U | GTTAGTATTATATTGGGGTAAGTTGTTTG | CCCTTCATCCCCAAACTCA | -510 to -371 | 140 | ||
| NT_006576 | SKP2 | m | GTCGTAGCGTCGTTCGTT | CTACAACCCGCTCTACTTCG | -198 to 10 | 208 |
| U | TTTTTTAGTTAGTTGTAGTGTTGTTTGTT | ACCCACTCTACTTCACAACCAC | -209 to 5 | 214 | ||
| NT_077569 | SPF45 | m | AGTGTCGTTCGGTTTCGTT | CCTCGAAAACTCCGACTACG | -216 to -6 | 211 |
| U | GTGGAGTGTTGTTTGGTTTTGT | AACTTACATCTAACACCTCCCAAA | -220 to -67 | 154 | ||
| NT_022517 | TOP2B | m | TGGGTTTCGTCGTTTCGT | CCGCGCTAAACCCGAAC | -241 to -66 | 176 |
| U | GGTTGTTGGGTTTTGTTGTTTT | TTCTCCTCAACCACCACACTAA | -254 to -60 | 195 | ||
| NT_010718 | TP53 | m | CGGAGTCGAGAGTTCGTG | CCGAAAACACTTTACGTTCG | 157 to 279 | 123 |
| U | GTTGAAAATATATGGAGTTGAGAGTTT | CTTTCCACAACAATAACACACTTC | 190 to 291 | 102 | ||
| NT_033903 | GSTP1 | m | GCGATTTCGGGGATTTTA | ACGACGACGAAACTCCAA | -183 to 16 | 199 |
| U | GTTGGGGATTTGGGAAAG | TATAAAAATAATCCCACCCCACT | -230 to -28 | 203 |
The methylation status of the promoter CpG islands of the 14 genes in all sample DNAs were analyzed by MSP on the sodium-bisulfite converted DNA[16,17]. The primer pairs for MSP and the genes’ genome access on GenBank are detailed in Table 3. In detail, 2 μg DNA in 50 μL TE was incubated with 5.5 μL of 3 mol/L NaOH at 37 °C for 10 min, followed by a 16 h treatment at 50 °C after adding 30 μL of fresh-prepared 10 mmol/L hydroquinone and 520 μL of freshly prepared 3.6 mol/L sodium-bisulfite at pH 5.0. The DNA was desalted using a home-made dialysis system with 10 g/L agarose. Then, the DNA (approximately 100 μL) was denatured at 37 °C for 15 min with 5.5 μL of 3 mol/L NaOH, followed by ethanol precipitation with 33 μL 10 mol/L NH4OAc and 300 μL ethanol. After washing with 700 mL/L ethanol, the gently dried DNA pellet was dissolved with 30 μL TE at 65 °C for 10 min. The DNA sample was finally stored at - 20 °C until further use. PCR reaction was carried out in a volume of 15 μL with 50 ng or less template DNA with JumpStart Taq polymerase (Sigma, USA). An initial denaturation step at 94 °C for 4 min was followed by 35 cycles of reaction at 94 °C for 20 s, variable annealing temperature (Table 3) for 20 s and 72 °C for 25 s, and a final extension at 72 °C for 5 min. The PCR products were separated by 12 g/L ethidium bromide containing agarose gel electrophoresis with 1 × Tris Acetate EDTA (TAE) buffer and visualized under UV illumination. To verify the PCR results, representative bands from each target were gel-purified and cloned into T-vector (Buocai, Shanghai, China), followed by automatic DNA sequencing provided by Buocai. Only verified results are presented in this report.
The M. Sss I treated DNA from the normal liver tissue was used in the MSP procedure as the control template. The DNA from the liver tissue of the healthy liver donor[14,16] was batch cleaved with EcoRI, followed by M. Sss I treatment overnight according to the manufacturer’s instruction (New England Biol., Boston, USA). The purified DNA was bisulphate treated as usual, and subjected to MSP with the primer pairs for 11 genes where no methylated alleles were detected in both cell lines and HCC tissues.
The established cell lines, BEL-7402 (human hepatocellular carcinoma cell line, No. TCHu68, Cell Bank in Shanghai), SMMC-7721 (human hepatocellular carcinoma cell line, No. TCHu13, Cell Bank in Shanghai), Hep3B (human hepatocellular carcinoma cell line, ATCC Number: HB-8064), HepG2 (human hepatocellular carcinoma cell line, ATCC Number: HB-8065), HCCLM3 (human hepatocellular carcinoma cell line, Cell Bank in Zhongshan Hospital, Shanghai), and L02 (immortalized normal liver cell line, No GNHu6, Cell Bank in Shanghai) were cultured in DEME medium plus 100 mL/L new born calf serum at 37 °C in a 50 mL/L CO2 atmosphere.
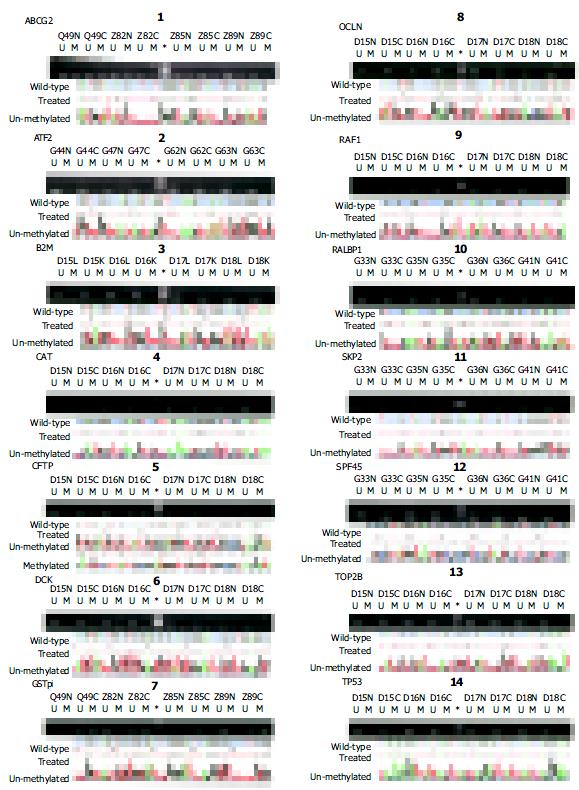
Total RNA from various HCC cell lines was extracted using the TriPure isolation reagent kit (Roche, USA). Reverse transcription was subsequently carried out using the superscript II RNase H-reverse transcriptase kit. β -actin was used as an internal control in separate reaction. The GSTpi cDNA primers (sense: 5’-GGAGACCTCACCCTGTACCA-3’; anti-sense: 5’-GGGCAGTGCCTTCACATAGT-3’) were the same as in a previously published paper[19]. The primer sequences and reaction conditions are listed in Figure 1. RT-PCR products were visualized under UV illumination after electrophoresis in 1.2% agarose, followed by the densitometric quantification.
The methylation data were dichotomized as 1 for the co-existence of the methylated and unmethylated alleles; 2, for methylated allele only; and 0 for the unmethylated for both alleles to facilitate statistical analysis using contingency tables. The statistic analyses for the association between the methylation profile of the gene and each of the clinical-pathological parameters were carried out with the statistics package (http://www.R-project.org/). Both Pearson’s Chi-square test with Upton’s adjustment and Fisher’s exact test (http://www.R-project.org/) were used to deal with the sample cells with the low expected values. The relative frequency with a 95% confidence interval (P < 0.05) for a binomial distribution was calculated for the tumors and the paired non-cancerous tissues.
Our previous efforts[14,16] provided an HCC specific altered pattern in DNA methylation of 11 among 44 genes by MSP in conjunction with sequencing verification. Among that list, there were several genes that might be classified as the drug resistance related genes, i.e., the hMLH1 and MGMT genes. The former maintained the unmethylated status and the later displayed hypermethylation in liver cancer. Although the impart of the tumor specific changes in methylation pattern of the MGMT gene has not been fully evaluated experimentally, it remains desirable to establish the methylation profile of more genes in the category of “drug resistance”. Such efforts should provide new insights into the underlying mechanisms as well as the capability of predicting HCC patient’s intrinsic profile of drug resistance, based upon the changes in DNA methylation pattern of the genes involved.
Sixty three genes were revealed from NCBI LocusLink database from the searching with “drug-resistance” as the key words (http://www.ncbi.nlm.nih.gov/LocusLink/list.cgi). To identify the genes falling into the category of CpG island containing genes, the sequence of the coding region plus 5 kb each upstream and downstream was downloaded and subjected to the analysis with CpG island identification software (http://http://www.ebi.ac.uk/emboss/cpgplot/). Forty four genes were found containing one or more CpG islands, but only 36 genes really had the promoter embedded in a CpG island (Table 1). Hence, transcription of these 36 genes is likely to be subjected to the control of the DNA methylation status. From these genes, 14 genes representing different functions and mechanisms in “drug-resistance” (Table 2) were selected for methylation profiling; three genes encoding the various membrane transporters: ABCG2, CFTR, and RALBP1, two encoding the transcription factors, ATF2, and TP53, four encoding the enzymes mediating the drug activation or inactivation: CAT, GSTpi, TOP2B and DCK, two encoding the proteins involved in signaling or cell-cycle progression: RAF1 and SKP2, and three with unknown underlying mechanisms: B2M, OCLN and SPF45.
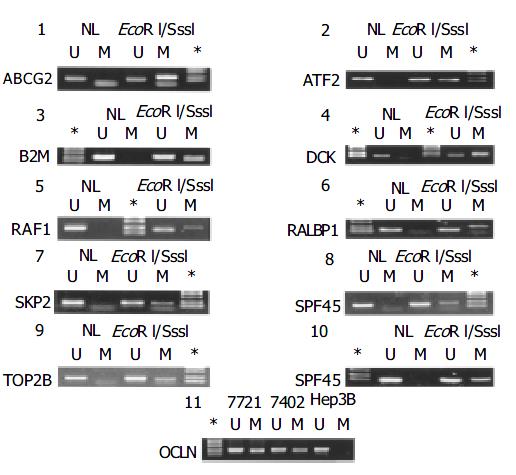
MSP is the most popular method for methylation profiling of any given targets in human cancers for both its easiness and sensitiveness. Based upon the distinguished difference in sensitivity of the methyl-cytosine from the cytosine to be converted to uridine by bisulphate treatment: the later is amenable, but the former is resistant, the methylation status of the target fragments can be determined by parallel PCR reactions with one pair of the primers in which the CG in the wide-type sequence is maintained for detection of methylated allele, and the other in which the CG is replaced with TG for the unmethylated allele. However, inherent with the PCR-type assays, there are compelling reasons to eliminate both false positive and negative PCR reactions. As 13 genes in this list except for the GSTpi gene had not been subjected to MSP previously, we carried out optimization of the PCR reaction with a panel of five established cell lines. For those targets, only the unmethylated allele was detected, MSP with the DNA from the normal liver tissue was carried out, showing that all 14 genes were unmethylated. To ensure that failure to detect the methylated allele was not false negative artifact in PCR reaction, we in vitro methylated the DNA from the normal liver with M. Sss I methyl-transferase that was capable of transferring methyl to the fifth carbon atom of the cytosine in the CpG dinucleotides, followed by methylation profiling. As shown in Figure 2, while no methylated allele was detected with the methylation targeted primers with the liver DNA, the M. Sss I treated DNA led to the positive PCR reaction, indicating that the PCR condition used for the tumor samples was capable of detecting the methylated targets, including ABCG2, ATF2, B2M, DCK, RAF1, RALBP1, SPF45, SKP2, TP53, and TOP2B genes (panels 1-10, respectively). As the rest four genes were heterozygously methylated in some of liver cancer cell lines, it indicated that the PCR reactions specific to the methylated allele was acceptable (for instance, lane 11 for OCLN, Figure 2), the in vitro methylation by M. Sss I testing was exempted. To eliminate the false negative results, the representative PCR bands were subjected to T-cloning and sequencing verification. Only the data, the bands of which had the correct identity in sequence, were regarded informative.
By MSP analysis, 13 genes displayed a uniform unmethylated status in the liver tissues from all four healthy donors, while the RAF1 gene was heterozygous in one case, but fully unmethylated in the rest three cases. In all 30 cases of HCC, the following 11 genes: ABCG2, ATF2, B2M, DCK, RAF1, RALBP1, SPF45, SKP2, TOP2B, OCLN and TP53 maintained the same unmethylated status in both HCC tissues and the paired non-cancerous tissues (Figure 3), indicating no DNA methylation mediated changes in the control of the expression of these 11 genes in HCC. The remaining three genes displayed in varying degree hypermethylation in HCC tissues (Figure 3 and Figure 1). Since hypermethylation of the CAT gene rarely occurred (3.3%) (Figure 3), its impacts on the HCC pathology might be rather trivial. Both GSTpi and CFTR genes were prevalently hypermethylated in HCC and their neighboring non-cancerous tissues (80% and 56.7%, and 77% and 50%, respectively, Figure 1), highlighting the possibility that inactivation of transcription of these two genes may indeed occur in HCC, and have certain etiological significance.
It has been well recognized that the so-called non-cancerous cells pathologically defined may have already suffered from certain genetic lesions as the corresponding cancerous tissues. Inclusion of the neighboring non-cancerous tissues in this study made it possible to analyze our results from the stage-specific perspective of carcinogenesis. As shown in Table 4, no significant difference was detected in the occurrence of hypermethylated GSTpi and CFTR genes between HCC and the paired non-cancerous tissues (P > 0.05) (Figure 1). If the neighboring non-cancerous tissues have indeed suffered from the early stage genetic and/or epigenetic lesions, the methylation changes of these two genes might occur at the early stage of HCC carcinogenesis.
| Gene | C/M | N/M | P |
| GSTpi | 24/30 | 17/30 | 0.052 |
| CFTR | 23/30 | 16/30 | 0.058 |
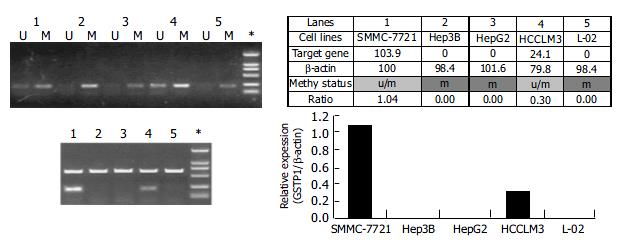
To correlate the hypermethylated status with the transcription silencing of GSTpi gene, we carried out MSP analysis of this gene and semi-quantitative RT-PCR for its expression in five established liver cancer cell lines. As shown in Figure 4, three liver cancer cell lines (i.e. HepG2, Hep3B and L-02) were fully methylated, and did not express GSTpi, while SMMC-7721 and HCCLM3 cell lines were heterozygously methylated, and expressed a detectable level of GSTpi mRNA. Therefore, the general notion stands correct in this case, that the hypermethylated status of the promoter CpG island is inversely correlated with the long-term transcription silencing state of the gene.
Glutathione-S-transferases (GSTs) are a family of phase II detoxification enzymes that catalyze the conjugation of glutathione to a wide variety of endogenous and exogenous electrophilic compounds, including subclasses of carcinogens and cytotoxic therapeutic drugs[20,21], so that the cells are protected from DNA damages[22]. Furthermore, the GSTpi isoform also regulates the mitogen-activated protein (MAP) kinase pathway that participates in cellular survival and death signals via protein: protein interactions with c-Jun N-terminal kinase 1 (JNK1) and apoptosis signal-regulating kinase (ASK1)[23]. Therefore, GSTs serve two distinct roles in the development of drug resistance to both substrate-type and other types of chemotherapeutic drugs via direct detoxification as well as acting as inhibitors of the MAP kinase pathway[24].
The cystic fibrosis trans-membrane conductance regulator (CFTR) belongs to the ABC transporter superfamily, and regulates the chloride permeability mainly in epithelial cells. Its defects may contribute to cystic fibrosis, a common autosomal recessive disorder among the Caucasian population, which affects multiple organs, including lung, pancreas, liver, sweat gland, reproductive system and intestine. CFTR gene has high expression level of transcripts in epithelial cells in pancreas and nasal polyps; low in lung, liver, and kidney; and undetectable in brain and fibroblasts[25]. It functions as a cAMP-regulated chloride channel, a conductance regulator by an autocrine mechanism involving ATP and/or chloride efflux, and an inhibitor of epithelial Na+ channels when chlorides are released[26]. The involvement of CFTR gene in drug resistance is implicated by observation that the forced over-expression of CFTR gene confers the multiple drug resistance to NIH 3T3 cells[27], a similar behavior to the founding member of this family, multiple drug resistance 1 (MDR1), which was identified three decades ago, for its etiological role in the cellular resistance to the multiple chemotherapeutic drugs. The unmethylated status of CFTR gene in the normal liver tissues suggested that its expression may be necessary for the physiological activities of hepatocytes. Although whether this gene is expressed in HCC has not been tested, the possibility does remain for the functional loss of this gene in HCC, indicated by the prevalent hypermethylation of its promoter CpG islands.
Therefore, lacking of expression of these two genes, implicated by the hypermethylated state of the promoter CpG islands, may make the cells more susceptible to the carcinogenic insults, a feature associated with the proneness to the malignant state of cells. Hypermethylation of the promoter CpG islands may reflect the long-term silencing state of transcription of both GSTpi and CFTR genes in HCC, which may offer growth advantages of the malignant cells over their normal counterparts in vivo. Indeed, the hypermethylation of both genes was an early phase event, as no statistically significant difference in the hypermethylation frequency between the paired neighboring non-cancerous and HCC tissues was observed (Table 4). Nevertheless, the hypermethylated status of these two genes may not be all bad, as the HCC with the hypermethylated alleles of these two genes (if either or both genes did not express) may be more sensitive to the relevant chemotherapeutic drugs than their unmethylated counterparts. Indeed, in a recent report, a better response to chemotherapy was achieved in breast cancer patients lacking GSTpi expression than the expressing counterparts[28].
Drug resistance of cancers represents a formidable challenge in cancer management, and the underlying mechanisms of its formation remain poorly defined. Both genetic and epigenetic defects can result in changes in expression of the “drug-resistance” genes in tumors. DNA methylation state of the promoter CpG islands has been demonstrated as a useful indicator for the transcriptional silencing state of the relevant genes. In this study, we determined the methylation profile of the genes at the focal point, classified as the drug-resistance type in DNA database in HCC, one of most devastating human cancers in Far East Asia, including China.
To identify suitable targets for such a study, we carried out the search with key word “drug resistance” from the LocusLink part of the human genome database. Among 63 entries (http://http://www.ncbi.nlm.nih.gov/locuslink/list.cgi), 44 (approximately 70%) fall into the category “the CpG island containing genes”, of which, 36 genes have their promoter embedded in the CpG island (Tables 2, 3). Hence, the DNA methylation mediated mechanism likely exerts the important control of over expression of this type of genes, the key function of which is somehow linked to the drug-resistance of cancer cells. Finally, 14 genes were selected for the methylation profiling in HCC. Although 11 genes maintained the unmethylated state in all the tissue samples tested, including four cases of the liver tissues from the healthy liver donors, 30 HCC tissues and their neighboring non-cancerous tissues, three genes, CAT, GSTpi and CFTR, exhibited to various degrees the hypermethylation status in HCC. CAT was only hypermethylated in one (3.3%) case. Both GSTpi and CFTR genes were prevalently hypermethylated (Figure 3 and Figure 1) in both HCC and the paired non-cancerous tissues. There was no significant difference in occurrence of the hypermethylated status between the HCC and the paired non-cancerous tissues, indicating that the hypermethylation of these two genes is likely to be the early rather than the late phase events behind the same rationale described previously[14]. This observation would strengthen the notion that the DNA methylation mediated transcription silencing of these two genes may promote carcinogenesis of HCC, by failing to protect the cells from both intracellular and extracellular genotoxic insults. On the other hand, this may offer certain advantages from the angle of the chemotherapeutic treatment of HCC. It is probably true that the HCC with hypermethylated GSTpi and CFTR genes may respond to the relevant chemotherapies better than the unmethylated counterparts. To further evaluate this hypothesis, both retrospective and prospective clinical studies specifically addressing the association between the chemotherapeutic responses of the HCC and the methylation state of the promoter CpG islands of the drug resistance genes, including GSTpi, CFTR and MGMT[14] genes with a larger cohort of HCC patients are underway.
Thanks are due to Jian-Guo Chen from Qidong Liver Cancer Institute, China, Li-Sheng Zhang from Guangxi Cancer Institute, Guangxi, China, Meng-Chao Wu from Eastern Hepatobiliary Surgery Hospital, Shanghai, China and Su-Shen Zhen, The First Affiliated Hospital, College of Medicine, Zhejiang University, Zhejiang, China for providing the patients’ samples.
| 1. | Parkin DM, Pisani P, Ferlay J. Global cancer statistics. CA Cancer J Clin. 1999;49:33-64, 1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1285] [Cited by in RCA: 1257] [Article Influence: 46.6] [Reference Citation Analysis (1)] |
| 2. | Bosch F. Global epidemiology of hepatocellular carcinoma. New York: Churchill Livingstone 1997; 13-28. |
| 3. | Bosch F; Aflatoxins. IARC Monogr Eval Carcinog Risks Hum. 1993;56:245-395. [PubMed] |
| 4. | Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of eighteen major cancers in 1985. Int J Cancer. 1993;54:594-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1065] [Cited by in RCA: 1042] [Article Influence: 31.6] [Reference Citation Analysis (1)] |
| 5. | Kruh GD. Introduction to resistance to anticancer agents. Oncogene. 2003;22:7262-7264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 67] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 6. | Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245-254. [PubMed] |
| 7. | Jones PA. Epigenetics in carcinogenesis and cancer prevention. Ann N Y Acad Sci. 2003;983:213-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 65] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 8. | Feinberg AP. Cancer epigenetics takes center stage. Proc Natl Acad Sci U S A. 2001;98:392-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 108] [Article Influence: 4.3] [Reference Citation Analysis (1)] |
| 9. | Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1012] [Cited by in RCA: 960] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 10. | Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1147] [Cited by in RCA: 1107] [Article Influence: 48.1] [Reference Citation Analysis (1)] |
| 11. | Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 694] [Cited by in RCA: 666] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 12. | Cho B, Lee H, Jeong S, Bang YJ, Lee HJ, Hwang KS, Kim HY, Lee YS, Kang GH, Jeoung DI. Promoter hypomethylation of a novel cancer/testis antigen gene CAGE is correlated with its aberrant expression and is seen in premalignant stage of gastric carcinoma. Biochem Biophys Res Commun. 2003;307:52-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 102] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 13. | Cao L, Owsianik G, Jaspers M, Janssens A, Cuppens H, Cassiman JJ, Nilius B. Functional analysis of CFTR chloride channel activity in cells with elevated MDR1 expression. Biochem Biophys Res Commun. 2003;304:248-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 14. | Yu J, Zhang HY, Ma ZZ, Lu W, Wang YF, Zhu JD. Methylation profiling of twenty four genes and the concordant methylation behaviours of nineteen genes that may contribute to hepatocellular carcinogenesis. Cell Res. 2003;13:319-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 82] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 15. | Burger P, Scheithauer B, Paulus W, Giannini C, Kleihues P. Astrocytic Tumours. In Pathology and Genetics of Tumours of the Nervous. Lyon: IARC Press. 2000;9-54. |
| 16. | Yu J, Ni M, Xu J, Zhang H, Gao B, Gu J, Chen J, Zhang L, Wu M, Zhen S. Methylation profiling of twenty promoter-CpG islands of genes which may contribute to hepatocellular carcinogenesis. BMC Cancer. 2002;2:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 136] [Article Influence: 5.7] [Reference Citation Analysis (1)] |
| 17. | Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990-2997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1369] [Cited by in RCA: 1391] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 18. | Cai L, Zhu JD. The tumor-selective over-expression of the human Hsp70 gene is attributed to the aberrant controls at both initiation and elongation levels of transcription. Cell Res. 2003;13:93-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 19. | Bakker J, Lin X, Nelson WG. Methyl-CpG binding domain protein 2 represses transcription from hypermethylated pi-class glutathione S-transferase gene promoters in hepatocellular carcinoma cells. J Biol Chem. 2002;277:22573-22580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 86] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 20. | Henderson CJ, McLaren AW, Moffat GJ, Bacon EJ, Wolf CR. Pi-class glutathione S-transferase: regulation and function. Chem Biol Interact. 1998;111-112:69-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 84] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 21. | Zimniak P, Nanduri B, Pikuła S, Bandorowicz-Pikuła J, Singhal SS, Srivastava SK, Awasthi S, Awasthi YC. Naturally occurring human glutathione S-transferase GSTP1-1 isoforms with isoleucine and valine in position 104 differ in enzymic properties. Eur J Biochem. 1994;224:893-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 327] [Cited by in RCA: 324] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 22. | Ryberg D, Skaug V, Hewer A, Phillips DH, Harries LW, Wolf CR, Ogreid D, Ulvik A, Vu P, Haugen A. Genotypes of glutathione transferase M1 and P1 and their significance for lung DNA adduct levels and cancer risk. Carcinogenesis. 1997;18:1285-1289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 260] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 23. | Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003;22:7369-7375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 995] [Cited by in RCA: 951] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 24. | Tew KD. Glutathione-associated enzymes in anticancer drug resistance. Cancer Res. 1994;54:4313-4320. [PubMed] |
| 25. | Bannykh SI, Bannykh GI, Fish KN, Moyer BD, Riordan JR, Balch WE. Traffic pattern of cystic fibrosis transmembrane regulator through the early exocytic pathway. Traffic. 2000;1:852-870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 26. | Morales MM, Capella MA, Lopes AG. Structure and function of the cystic fibrosis transmembrane conductance regulator. Braz J Med Biol Res. 1999;32:1021-1028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 27. | Wei LY, Stutts MJ, Hoffman MM, Roepe PD. Overexpression of the cystic fibrosis transmembrane conductance regulator in NIH 3T3 cells lowers membrane potential and intracellular pH and confers a multidrug resistance phenotype. Biophys J. 1995;69:883-895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 28. | Su F, Hu X, Jia W, Gong C, Song E, Hamar P. Glutathion S transferase pi indicates chemotherapy resistance in breast cancer. J Surg Res. 2003;113:102-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
Co-first-authors: Sheng Ding, Bang-Dong Gong and Jian Yu
Edited by Xia HHX Proofread by Zhu LH and Xu FM