Published online Nov 1, 2004. doi: 10.3748/wjg.v10.i21.3188
Revised: February 19, 2004
Accepted: February 26, 2004
Published online: November 1, 2004
AIM: To construct a yeast expression system of human augmenter of liver regeneration (hALR) and to examine its bioactivity in vitro.
METHODS: With PCR and gene recombination techniques, cDNA of open reading frame of hALR was obtained from recombinant plasmid pcDNA3.1-hALR and inserted into plasmid pPIC9. The cDNA of hALR from recombinant plasmid pPIC9-hALR demonstrated by sequencing was subcloned into plasmid pPIC9K. The recombinant plasmid pPIC9K-hALR was transformed into GS115 with electroporation. hALR was expressed by GS115 under the induction of 5 mL/L methanol and purified with ultrafiltration after it was analyzed by 15% SDS-PAGE and Western blot. The effects of hALR on in vitro proliferation of QGY and HepG2 cells were evaluated by 3H-TdR methods.
RESULTS: The correctness and integrity of recombinant plasmids pPIC9-hALR and pPIC9K-hALR were identified by restriction digestion, PCR and sequencing methods, respectively. hALR as a secretive protein was successfully expressed by GS115. Its molecular weight was about 15 ku and the target protein was about 60% of the total protein in the supernatant from GS115 with plasmid pPIC9K-hALR. The results of Western blot of hALR showed the specific band. The high qualitative hALR was obtained through ultrafiltration. hALR could stimulate in vitro proliferation of QGY and HepG2 cells in a dose-dependent manner, but there was a difference in reactivity to hALR between QGY and HepG2.
CONCLUSION: The hALR as a secretive protein can be successfully expressed by GS115. It may stimulate in vitro proliferation of QGY and HepG2 cells at a dose-dependent manner. But QGY and HepG2 cells have different reactivities to hALR.
-
Citation: Liu Q, Yu HF, Sun H, Ma HF. Expression of human augmenter of liver regeneration in pichia pastoris yeast and its bioactivity
in vitro . World J Gastroenterol 2004; 10(21): 3188-3190 - URL: https://www.wjgnet.com/1007-9327/full/v10/i21/3188.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i21.3188
Augmenter of liver regeneration (ALR) is an important cytokine recently discovered and cloned from rat livers[1,2]. It could stimulate DNA synthesis of liver cancer cells in vitro and enhance liver regeneration in rats after partial hepatectomy, but there is a debate whether it could directly stimulate DNA synthesis of hepatocytes in vitro[1,3]. It has also been proved that ALR could partially reverse the liver fibrosis in experimental rats[4] and enhance the success rate of fetal pancreas transplantation in rats[5]. It seems to bring a bright future in the treatment of severe liver diseases. Although the recombinant human and rat ALR were successfully expressed in E.coli in our laboratory[6-8], there are some defects that the target protein as an inclusion body expressed in E.coli could not often be refolded and modified correctly and that the procedure to purify ALR is very complicated. Because the pichia pastoris expressing system could overcome all these defects, it was selected to express ALR, which could be secreted into supernatants so that it was easy to purify ALR. It is possible for us to carry out further study with purified ALR.
Escherichia coli JM109 and TOP10F’, plasmid pcDNA3.1-hALR, hepatocarcinoma HepG2 and QGY strains were preserved and monoclonal antibody against hALR was made in our laboratory[9]. Pitch pastoris GS115, plasmids pPIC9 and pPIC9K were from Invitrogen Corporation, USA. DNA ladder marker, restriction endonuclease, and T4 DNA ligase were from Shanghai Sango, China. Peptone and YNB were from Amresco Co. USA. Yeast power was from Oxoid. 8200 cell filter, 5 kD and 30 kD membranes were from Millipore Co. USA. Gene pulser-II was from Bio-Rad Co. Ltd., USA.
According to the maps of plasmids pcDNA3.1-hALR, pPIC9 and pPIC9K, the sequences of primers were designed as follows: forward:P1 5’-CACTCGAGAAAAGACGGACCCAGCAGAAG C-3’.reverse:P25’-CTGCGGCCGCTTAGTCACAGGAGCCGTC CTTCC-3’. XhoI and NotI restriction sites were introduced and defined by the underline. Primers were synthesized by BoYa Co. Ltd. hALR sequence was amplified by polymerase chain reaction (PCR) for 30 cycles from pcDNA3.1-hALR containing the full-length open reading frame of hALR gene. Each cycle consisted of denaturation for 45 s at 94 °C, annealing for 45 s at 62 °C, and extension for 1 min at 72 °C. The PCR products were analyzed on 15 g/L agarose gel electrophoresis as specific band examination.
The PCR products of hALR from the gel extraction and pPIC9 were digested with Xho I and Not I and then purified, respectively. pPIC9 and hALR fragments were mixed with 3:1 mol/L and ligated by T4 DNA ligase overnight at 16 °C, then transformed into JM109 with CaCl2 method. The positive clones were selected in LB plates with ampenicillin and identified by PCR, restriction enzyme digestion and sequencing, respectively. After the correction and integrity of recombinant pPIC9-hALR were proved, pPIC9-hALR and pPIC9K were digested with SalI and SacI. A 3.4 kb fragment containing hALR was obtained from pPIC9-hALR and 6.3 kb fragment from pPIC9K. Both fragments were mixed with 2:1 mol/L, ligated by T4 DNA ligase and transformed into TOP10F’ with CaCl2 method. The positive clones were selected in LB plates with ampenicillin and karamycin and the plamids from them were identified by PCR and restriction enzyme digestion on the next day.
Recombinant plasmids pPIC9K-hALR (8 μL) and pPIC9K (8 μL) were linearized, respectively, by digestion with SalI. Competent pitch pastoris GS115 was prepared according to the manual of Invitrogen Corporation. The linear palsmids pPIC9K (5-10 μg) and pPIC9K-hALR (5-10 μg) were mixed with competent GS115 (80 μL), respectively. Under the conditions of 1500V, 25 μF and 200 Ω , the competent GS115 was transformed by different mixtures using the Bio-Rad gene pulser, then planted and cultured in plates of minimal dextrose medium without histidine at 30 °C for 2 d for screening positive clones. The positive clones with pPIC9K-hALRp or PIC9K were respectively cultured in 3 mL buffered glycerol-complex medium (BMGY) overnight after identified by PCR. Then 15 μL of GS115 was planted into 5 mL BMGY medium and cultured at 28 °C until the optical density at 600 nm (A600) reached four. The GS115 with pPIC9K-hALRp and PIC9K were collected by centrifugation, resuspended by buffered methanol-comlex medium to A600 reaching one and cultured continuously at 28 °C for 3 d. The 5 mL/L methanol in the medium was kept by methanol compensation at every 24 h. The expressed products at the different time points were analysed by 160 g/L SDS-PAGE and the expressed level was evaluated by computer scan.
Supernatants from GS115 with pPIC9K and pPIC9K-hALR were respectively loaded on 150 g/L SDS-PAGE for protein separation and transferred to polyvinylidene difluoride membrane by electrotransfer. After blocked with 50 mL/L milk in PBS, the membrane was incubated with monoclonal antibody (diluted 1:2 000) against hALR for 1 h at room temperature, washed three times with phosphate buffered saline, then mixed with goat anti-mouse IgG antibody coupled with horseradish peroxidase for 45 min, the bands were visualized by 3-3’-diaminobenzidine (DAB).
The ultrafiltration purification was selected according to the SDS-PAGE analysis of the supernatant from GS115 with pPIC9K-hALR there were no other bands except target one under 50 kD. The supernatant from GS115 induced by pPIC9K-hALR was centrifuged at 3 000 r/min for 10 min, and passed through 30 kD and 5 kD membranes and the filtrate between 30 kD and 5 kD was collected. As a control the supernatant from GS115 with pPIC9K was treated with same procedure as described above. The purified products were analyzed by 160 g/L SDS-PAGE and qualified by Lowary methods.
Human hepatoma cell lines HepG2 and QGY were planted at a concentration of 1 × 104cell/well in 96-well plates supplemented with 80 mL/L fetal bovine serum, 5 × 10-5 mol/L penicillin, 80 μg/mL streptomycin, and stimulated with different doses of hALR expressed by yeast and E.coli at 37 °C, 50 mL/L CO2 for 8-9 h. The supernatant from GS115 with pPIC9k as negative control was also added. Three parallel wells were stimulated by the same concentration of hALR. The cells were harvested after 12 h of culture by adding 1 μCi /well 3H-TdR. The count per minute (cpm) value of cells from each well was measured by liquid scintillation counter.
The data were expressed as mean ± SD. Comparison was carried out using analysis of variance. P value less than 0.05 was considered statistically significant.
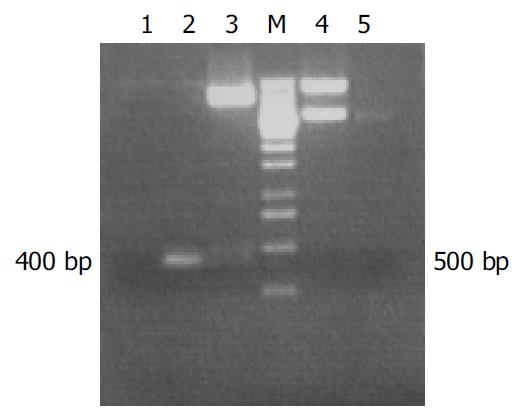
The specific 400 bp fragment of hALR amplified by PCR from plasmid pcDNA3.1-hALR was analysed by 15 g/L agarose gel electrophoresis. The correctness and integrity of hALR fragments in recombinant plasmid pPIC9-hALR were proved by XhoI + NotI digestion and sequencing. The 3.4 kb fragment containing hALR was released from pPIC9K-hALR by digestion with salI and sacI. The 400 bp fragment was also obtained from pPIC9K-hALR by PCR (Figure 1).
Electrophoresis of supernatants from GS115 induced by pPIC9K-hALR showed that there was a specific band with 1.5 × 104 dalton molecular weight coinciding with the theoretical value and the target protein was about 60% of the total protein in the supernatant. There was no corresponding band in supernatant from GS115 transfected with pPIC9K. A single band of hALR was obtained through ultrafiltration purification (Figure 2).
Western blot result showed that the supernatant from GS115 with pPIC9K-hALR could specially bind to monoclonal antibody against hALR in 15 kD site, but the supernatant from GS115 with pPIC9K could not bind (Figure 3).
Results showed that hALR from GS115 with pPIC9K-hALR could stimulate the proliferation of HepG2 and QGY on nanogram level in a dose-dependent manner. There was a marked difference in the reactivity to hALR between QGY and HepG2 although both cells were from human liver cancer. In addition, hALR from yeast had stronger effect on the proliferation of HepG2 and QGY than that from E.coli at same concentration (Table 1).
| Cells | Control | Concentrations of hALR from yeast and E.coli | |||||
| 0.1 ng | 10 ng | 1 μg | 8 μg | 16 μg | |||
| HepG2 | 7 174 ± 494 | yeast | 1 3779 ± 1 431a | 13 679 ± 1 205a | 15 279 ± 1 802a | 22 670 ± 1 402bc | 24 958 ± 1 326bc |
| E.coli | 1 0754 ± 2 756a | 11 189 ± 1 447a | 12 997 ± 1 683a | 13 858 ± 440a | 14 411 ± 1 452a | ||
| QGY | 821 ± 11 | yeast | 2 549 ± 526bc | 2 298 ± 203bc | 3 188 ± 564bc | 11 825 ± 841bc | 17 021 ± 795bc |
| E.coli | 934 ± 50a | 883 ± 106 | 1 222 ± 168a | 1 362 ± 171a | 6 209 ± 225b | ||
The Pichia pastoris system has been widely used in recent years due to its simple procedure, low expenditure and easy scaling up. It is more important that the foreign protein can be correctly refolded and modified, more than 200 proteins have been expressed by this system[10]. Our results showed that hALR with 1.5 × 104 dalton molecular weight as a secretive protein was successfully expressed by GS115 and about 60% of total protein was expressed in the supernatant. The ultrafiltration purification for hALR in the supernatant was applied because there were no other bands around the target protein, and a satisfactory purification effect was achieved. Our results demonstrated that not only the purification efficiency was greatly enhanced, but also the purification procedure was obviously simplified. Although it was reported that rat ALR could not directly stimulate liver cancer cells in vitro[1], our data demonstrated that hALR could stimulate QGY and HepG2 proliferation in a dose-dependent manner. Previously, we and others also obtained the similar results in rat ALR researches[11,12]. These results suggested that ALR enhanced liver regeneration not only through indirect pathway i.e. immune regulation[1,13-16], but also through direct one. Our result showed that at the same concentration, hALR expressed in yeast had a stronger effect on the proliferation of HepG2 and QGY than that expressed in E.coli, although it was reported that there was no N-glycosylation site in its amino acid sequence[1]. Our result suggests that there might exist some unknown modifications such as O-glycosylation site in hALR molecules expressed by yeast, and modification of protein after translation is very important for keeping higher bioactivity of target protein. Hence, the successful expression of ALR in pichia postoris and its purification may provide a good tool for further research in this field.
| 1. | Hagiya M, Francavilla A, Polimeno L, Ihara I, Sakai H, Seki T, Shimonishi M, Porter KA, Starzl TE. Cloning and sequence analysis of the rat augmenter of liver regeneration (ALR) gene: expression of biologically active recombinant ALR and demonstration of tissue distribution. Proc Natl Acad Sci USA. 1994;91:8142-8146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 143] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 2. | Hagiya M, Francavilla A, Polimeno L, Ihara I, Sakai H, Seki T, Shimonishi M, Porter KA, Starzl TE. Cloning and sequence analysis of the rat augmenter of liver regeneration (ALR) gene: expression of biologically active recombinant ALR and demonstration of tissue distribution. Proc Natl Acad Sci USA. 1995;92:3076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 3. | Yang XM, Xie L, Qiu ZH, Gong F, Wu ZZ, He FC. cDNA clone, expression and biological activity study of augmenter of liver regeneration. Shengwu Huaxue Zazhi. 1997;13:130-135. |
| 4. | Wang AM, Yang XM, Guo RF, Zhang L, Li PJ, Wang QM, He FC. The recombinant agumenter of liver regeneration reverse fibrosis in experimental rat. Zhonghua Ganzangbing Zazhi. 1999;7:243. |
| 5. | Adams GA, Maestri M, Squiers EC, Alfrey EJ, Starzl TE, Dafoe DC. Augmenter of liver regeneration enhances the success rate of fetal pancreas transplantation in rodents. Transplantation. 1998;65:32-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 6. | Liu Q, Wang Z, Luo Y. [The cDNA clone and sequence analysis of the coding region of human augmenter of liver regeneration (hALR) gene]. Zhonghua Ganzangbing Zazhi. 1999;7:156-158. [PubMed] |
| 7. | Liu Q, Shi X. [Construction of prokaryotic expression vector of hALR and its expression in E.Coli]. Zhonghua Ganzangbing Zazhi. 2000;8:9-11. [PubMed] |
| 8. | Ma HF, Liu Q. The cDNA clone of the coding region of rat augmenter of liver regeneration gene and its prokaryotic expression. Chongqing Yikedaxue Xuebao. 2002;27:9-11. |
| 9. | Ma HF, Liu Q. Preparation of monoclonal antibody to human augmenter of liver regeneration: screening of hybridomas with unpurified antigen expressed by E.coli. Zhongguo Mianyixue Zazhi. 2002;18:671-673. |
| 10. | Cregg JM, Cereghino JL, Shi J, Higgins DR. Recombinant protein expression in Pichia pastoris. Mol Biotechnol. 2000;16:23-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 629] [Cited by in RCA: 621] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 11. | Yu HF, Liu Q. [Expression of rat augmenter of liver regeneration in pichia pastoris and evaluation of its bioactivity in vitro]. Zhonghua Ganzangbing Zazhi. 2003;11:421-423. [PubMed] |
| 12. | Yang XM, Hu ZY, Xie L, Wu ZZ, Wu CT, He FC. [In vitro stimulation of HTC hepatoma cell growth by recombinant human augmenter of liver regeneration (ALR)]. Shengli Xuebao. 1997;49:557-561. [PubMed] |
| 13. | Francavilla A, Vujanovic NL, Polimeno L, Azzarone A, Iacobellis A, Deleo A, Hagiya M, Whiteside TL, Starzl TE. The in vivo effect of hepatotrophic factors augmenter of liver regeneration, hepatocyte growth factor, and insulin-like growth factor-II on liver natural killer cell functions. Hepatology. 1997;25:411-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 14. | Gandhi CR, Kuddus R, Subbotin VM, Prelich J, Murase N, Rao AS, Nalesnik MA, Watkins SC, DeLeo A, Trucco M. A fresh look at augmenter of liver regeneration in rats. Hepatology. 1999;29:1435-1445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 81] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Polimeno L, Margiotta M, Marangi L, Lisowsky T, Azzarone A, Ierardi E, Frassanito MA, Francavilla R, Francavilla A. Molecular mechanisms of augmenter of liver regeneration as immunoregulator: its effect on interferon-gamma expression in rat liver. Dig Liver Dis. 2000;32:217-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 16. | Tanigawa K, Sakaida I, Masuhara M, Hagiya M, Okita K. Augmenter of liver regeneration (ALR) may promote liver regeneration by reducing natural killer (NK) cell activity in human liver diseases. J Gastroenterol. 2000;35:112-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
Edited by Kumar M and Wang XL Proofread by Xu FM