Published online Nov 1, 2004. doi: 10.3748/wjg.v10.i21.3141
Revised: March 5, 2004
Accepted: March 12, 2004
Published online: November 1, 2004
AIM: To establish the transgenic mouse line harbouring complete hepatitis B virus (HBV) genome with mutant s gene (adr subtype).
METHODS: Transgenic mice were generated by microinjecting HBV genome into fertilized eggs. Integration, expression, replication of HBV gene and histological changes in transgenic mice were estimated by genomic DNA PCR, serum DNA PCR, Southern blot, ELISA, HE staining, immunohistochemistry and transmission electron microscopy. Transgenic mice with HBsAg positive in serum were bred and analyzed.
RESULTS: A total of 288 eggs survived from microinjections were transplanted into the oviducts of 13 pseudopregnant mice and 49 pups were produced. Twenty-six mice were identified to have the integrated HBV gene. Serum HBsAg and HBeAg were detected in 2 of 43 mice. HBsAg and HBcAg in cytoplasm or nuclei of hepatocytes were detected in 10 mice. Founders with HBsAg in serum were named lineages G145R-15 and G145R-18. Of the 16 F1 offsprings generated by G145R-15 founder, 12 were positive for HBV genome with PCR, 10 were positive for HBsAg and HBcAg with immunohistochemistry and 7 were positive for HBsAg and HBeAg with ELISA. Only 1 of 8 F1 offsprings generated by G145R-18 founder was survived and it was detected positive for HBV genome, HBsAg, HBcAg and HBeAg. Both of the two lineages had some pathological characteristics of mild chronic hepatitis B in the liver, such as swelling of hepatocytes and focal hepatocellular necrosis and parenchymal lymphomononuclear cell infiltrate.
CONCLUSION: Transgenic mice harbouring HBV with mutant s gene can be generated. The HBV genes are integrated in the transgenic mice genome and can be expressed, replicated, packaged and excreted. HBV DNA can be stably transmitted in the transgenic mice.
-
Citation: Ge JH, Zhang LZ, Li JX, Liu H, Liu HM, He J, Yao YC, Yang YJ, Yu HY, Hu YP. Replication and gene expression of mutant hepatitis B virus in a transgenic mouse containing the complete viral genome with mutant
s gene. World J Gastroenterol 2004; 10(21): 3141-3145 - URL: https://www.wjgnet.com/1007-9327/full/v10/i21/3141.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i21.3141
Hepatitis B virus (HBV) is a major pathogen causing human acute and chronic hepatitis B[1] and has a very close association with cirrhosis and human hepatocellular carcinoma (HCC)[2-5]. Hepatitis B (HB) vaccine can protect human from infecting HBV. Many children born from HBeAg positive mothers would suffer from HBV after HB vaccination because of the mutation of HBV[6-7]. Genetic mutation in viruses is usually associated with escape of host immune responses, development of drug resistance, modification of virulence and patterns of epidemiology of diseases. In addition, HBV has a limited host range, which impedes much of the study on its biological characteristics and human hepatitis B. Over the past several years, many transgenic lineages harbouring intact HBV gene[8] or expressing the HBV envelope, core, precore and X proteins[9] under the control of HBV or cellular liver-specific promoters have been generated, thereafter, the transgenic mouse system was applied in HBV studies[10-12]. However, no reports of transgenic mouse harbouring mutant HBV are available.
HBV belongs to hepadnavirus family, containing a small (3.2-kb), circular, double-stranded DNA genome. The minus strand includes at least four open reading frames (ORF), of which, S-ORF is divided into pres1, pres 2 and s gene. HBsAg encoded by the s gene is the major component of hepatitis B vaccine. It is also an important diagnostic evidence of HBV infection. Hepatitis B immunoglobulin could be used to protect the liver from reinfection after liver transplantation, but it depends on the interaction of HBsAg and anti-HBs. Therefore, the change of antigenicity of HBsAg has a direct impact on the diagnosis and therapy of HBV. It is most common to find the mutation at aa145 where glycine is substituted by arginine (G145R). This mutant could result in the failure of HB vaccination[13] and failing to protect the liver from reinfection after liver transplantation[14-16]. Recently, several lines of evidence of horizontal transmission have been found[17-18]. It brings about many new problems to prophylaxis and therapy of HB. It is necessary to study further on the change of biological characteristics related with this point mutation. Transgenic mice harbouring G145R mutant are a good animal model in study of changes of antigenicity and immunogenicity as well as pathogenicity of HBV. Therefore, we generated transgenic mice harboring complete genome of HBV (adr subtype) G145R mutant by microinjection method, in which HBsAg and HBeAg could be expressed, and genomic DNA of HBV could be replicated and packed into complete virus particles. This model makes it possible to examine many aspects of HBV and its associated biomedical issues in vivo, and is an animal model for drug screen and therapy involved in this mutant.
Restriction endonucleases and T4 DNA ligase were obtained from Sangon Co. Canada. Plasmid P II was endowed by Dr. Jian-Wen He and contained overlength HBV genomes, beginning at the middle of the X gene, terminating at nucleotides 1982, just downstream of the unique polyadenylation signal in the HBV genome at a unique BamHI site. Plasmid P II contained a point mutation at the site of s gene nt 587(G→A). HBsAg and HBeAg ELISA reagents were purchased from Abbott Laboratories and Sino-American Biotechnology Co., respectively. Mouse monoclonal antibody against HBsAg and rabbit HBc/eAg primary anti-serum were purchased from DAKO, USA. Sheep anti mouse IgG-HRP was obtained from CALBIOCHEM, Germany. Serum DNA extraction kit was got from Sino-American Biotechnology. QIA quick gene gel kit and plasmid extraction kit were purchased from QIA-gene. C57BL/6 mice were SPF level and maintained on a 14:10 light-dark schedule (lights off at 10 pm, lights on at 8 am every day).
HBV transgenic mice were produced by microinjection of the Pvu II fragment excised from plasmid P II into C57BL/6 embryos by conventional technology. In brief, the Pvu II fragment containing 1.2 copy HBV DNA was gel purified and dissolved in TE buffer (10 mmol/L Tris-HCl, 0.2 mmol/L EDTA, pH7.5) at a final concentration of 1 mg/L (2000 copies/pL). After it was injected into male pronuclei obtained from C57BL/6 females, the eggs were implanted into oviducts of pseudopregnant recipients to enable further development before term. Founder animals were screened by analysis of serum for HBsAg and HBeAg. Animals positive for both antigens were expanded by repetitive backcrossing against the C57BL/6 strain and bred for analysis.
To isolate the total genomic DNA, approximately one third of the tail of 10-d-old mice was cut and placed into a 1.5 mL microcentrifuge tube containing 500 μL of TB buffer. The tubes containing the tail fragments were incubated overnight at 55 °C. DNA was extracted once with 500 μL of 1:1 (v/v) equilibrated phenol-chloroform, and precipitated with 2 volumes of ethanol. After centrifugation, precipitates were resuspended in 500 μL water.
Total tail genomic DNA (1 mg) was analyzed by PCR using HBV-specific primers (5’-CCCAA CCTCC AATCA CTCAC CAACC-3’ [sense, nt 2 139 to 2 153] and 5’-GGCCC CCAAT ACCAC ATCAT CCATA -3’ [antisense, nt 2 583 to 2 556]). A 20 μL of samples of the products from direct PCR amplifications was analyzed by electrophoresis on a 10 g/L agarose gel in the presence of 0.5 μg of ethidium bromide per mL. DNA bands were visualized by UV fluorescence.
Southern blot analysis was performed on the total genomic DNA by agarose gel electrophoresis of 20 μg of restricted genomic DNA as previously described. Before electrophoresis, all DNA samples were digested with EcoR I at 10 U/μg for 4 h at 37 °C. Nylon filters were hybridized with HBV-specific digoxigeninlabeled DNA probes as previously described[19].
Total serum genomic DNA from transgenic mice was extracted according to the manufacturer’s instructions, and analyzed by PCR using HBV-specific primers as described above.
In selected experiments, liver tissue derived from transgenic mouse lineages was also studied. In some experiments, mice were anesthetized with pentobarbitone (Harvest Pharmaceutical Co. LTD) prior to phlebotomy from the retro-orbital plexus and before they were sacrificed by cervical dislocation. In the other experiments, mice were anesthetized with pentobarbitone prior to biopsy of the liver at the age of 3, 6, 9, 12, 15, 18 mo. Intracellular distributions of HBcAg and HBsAg were assessed by the labeled-avidin-biotin detection procedure[20]. Briefly, paraffin-embedded sections in PBS, pH7.4, were treated for 15 min at 37 °C with 30 mL/L hydrogen peroxide and washed with PBS. After the sections were blocked with normal goat serum for 30 min at room temperature, rabbit anti-HBc/eAg primary antiserum or mouse anti-HBsAg monoclonal antibody was applied at a 1:100 (HBcAg) or 1:500 (HBsAg) dilution for 12 h at 4 °C. After washed with PBS, a secondary antiserum consisting of biotin-conjugated goat anti-rabbit or goat anti-mouse immunoglobulin G was applied at a 1:100 dilution for 30 min at 37 °C. The antibody coated slides were washed with PBS, treated with streptavidin-horseradish peroxidase conjugates at a 1:600 dilution for 30 min at 37 °C, stained with nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl-phosphate (NBT/BCIP, MAXIN Co.), and counterstained with Mayer’s hematoxylin before mounted.
Transgenic mice of lineages G145R-15 and G145R-18 were anesthetized with pentobarbitone prior to phlebotomy from the retro-orbital plexus 200 μL every week. Serum was isolated by centrifugation. HBsAg and HBeAg concentrations were analyzed every month by using commercially available enzyme-linked immunosorbent assay reagents according to the manufacturer’s instructions.
Serum from transgenic mice was also assayed for anti-HBsAg, anti-HBeAg and anti-HBcAg every month by using commercially available enzyme-linked immunosorbent assay reagents according to the manufacturer’s instructions.
Intracellular nucleocapsid particles were analyzed exactly as described elsewhere. Briefly, thin liver sections were fixed overnight at 4 °C in 40 g/L paraformaldehyde-1 g/L glutaraldehyde in PBS. Sections were then postfixed in 10 g/L OsO4 in cacodylate buffer (pH7.4) for 1 h at room temperature, dehydrated in gradient ethanol, and embedded in epoxy resin (TAAB 812; Emmer Green, Reading, England). Sections were cut on a LKB Ultratome III, mounted on copper grids, stained in uranyl acetate and lead citrate, and viewed with a Hitachi H-800 electron microscope.
Serum-derived virus particles were analyzed as follows. Five hundred microliters of transgenic-mouse serum was centrifuged (300 000 g) for 12-16 h at 4 °C through 2.4 mL of 100 g/L sucrose into 0.8 mL of 600 g/L sucrose. Fractions (100 μL) were collected from the bottom, and the first fraction was incubated for 45 min at room temperature on MA18/7-coated (100 μg/mL) Parlodion-carbon-coated nickel grids that were subsequently drained with a piece of filter paper and washed twice in PBS. The grids were then fixed in 15 g/L glutaraldehyde for 2 to 5 min, washed twice in H2O, negatively stained with uranyl acetate, and viewed with a Hitachi H-800 electron microscope.
Tissue samples were fixed in 40 g/L neutral buffered formaldehyde, embedded in paraffin, sectioned (4 μm), and stained with hematoxylin and eosin as described[8].
Serum alanine aminotransferase (sALT), serum aspartate aminotransferase (sAST) , blood urea nitrogen (BUN), creatinine (Cr) and γ-glutamyl transpeptidase(γ-GT) were tested with an auto-biochemical analyzer.
Hundreds of molecules of target fragments were microinjected into male pronuclei of fertilized eggs. A total of 288 eggs survived from microinjections were transplanted into the oviducts of 13 pseudopregnant mice and consequently, the mice were pregnant and gave birth to 49 pups. In addition, 43 were survived. HBsAg and HBcAg in cytoplasm or nuclei of hepatocytes were detected in 10 mice. Two of 43 founder animals whose sera were positive for both HBsAg and HBeAg were produced and named lineages G145R-15 and G145R-18. They were expanded by repetitive backcrossing against the C57BL/6 strain and studied in detail. Sixteen F1 offsprings of lineage G145R-15 founder mice were born. Twelve of them were identified as the integration of HBV gene by PCR and Southern blot hybridization analysis of tissues. HBsAg and HBcAg were detected in eight of them by immunohistochemistry. HBsAg and HBeAg were detected in seven of them by using ELISA. Only one of eight F1 offsprings generated by lineage G145R-18 founder was survived. In addition, all above methods detected it positive. F1 offsprings were positive for both HBsAg and HBeAg and expanded by repetitive backcrossing against the nontransgenic C57BL/6.
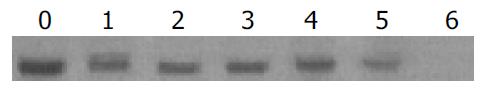
Total DNA was isolated from tail tissues of lineages G145R-15 and G145R-18, digested with an enzyme (EcoR I) that was not cut within the transgene, and analyzed by Southern blot (Figure 1).
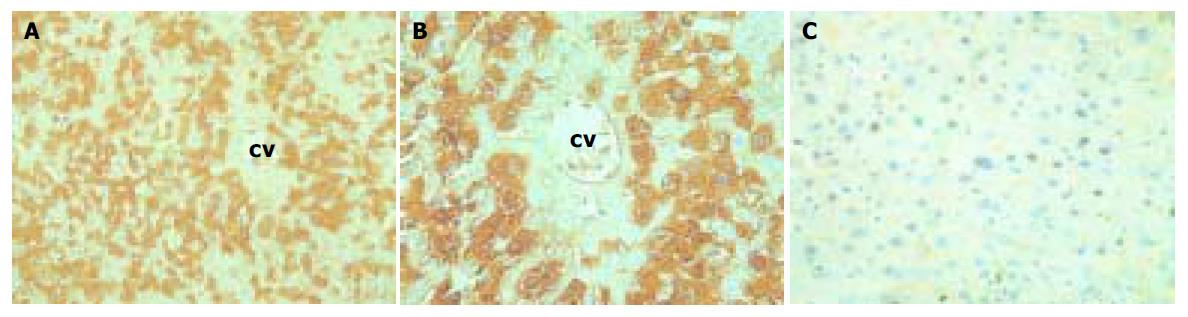
At the protein level, HBsAg (a product of the 2.1-kb mRNA) and HBeAg (a product of the 3.5-kb mRNA) were easily detectable in the serum in both lineages (Table 1). As expected, HBcAg (also a product of the 3.5-kb mRNA) and HBsAg were easily detectable in the liver by immunohistochemical staining (Figures 2A, B).
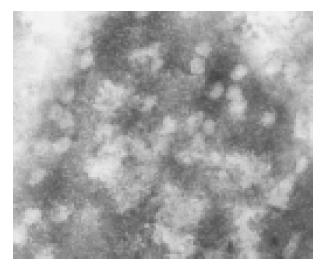
Both lineages displayed the same subcellular and architectural distribution of HBcAg in the liver. Figure 2A and 2B shows that HBcAg and HBsAg were present in the cytoplasm of almost all the hepatocytes distributed widely throughout the hepatic lobules from the centrilobular region around central veins to the periportal region around the portal tracts, but positive hepatocytes surrounding the central veins were much more than those around centrilobule. At the nucleic level, however, only HBcAg was detectable (Figure 2C). Typical 25-30 nm HBcAg particles (core particles and nucleocapsid particles) were detected in the nuclei (Figure 3). All indicated the existence of important host influences on viral gene expression in these animals.
To determine whether potentially infectious viral particles were formed and secreted into the blood of these mice, serum was analyzed for the presence of HBV DNA and sedimentable particles displaying the ultrastructural characteristics of complete virions (Dane particles). HBV DNA was detectable in the serum of an HBV transgenic-mouse lineage that replicated the viral genome.
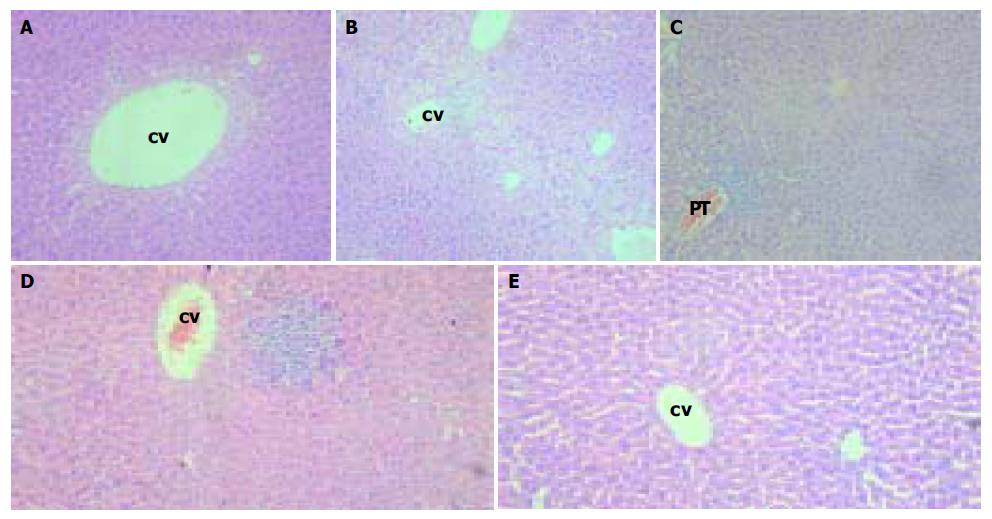
Animals from lineages G145R-15 and G145R-18 were monitored histologically for over eighteen months without evidence of pathological changes in the kidney, brain, spleen, lung, testis, ovaries, skin, and muscle except for the liver. In some region of the liver, swollen hepatocytes, hydropic degeneration or ballooning degeneration was observed in hepatic lobules that were distributed in the centrilobular region around central veins (Figure 4A, B). In the other, focal or unicellular necrosis, together with predominantly lymphocyte infiltrate was present (Figure 4D). In the portal or periportal tract, mild or moderate inflammation was also observed, as illustrated in Figure 4C. Interface inflammation could be observed in some cases. However, fibrosis was absent. Acidophilic body was scarce.
The ELISA test indicated that there was no significant positive result of HBV antibodies in HBV transgenic mouse serum.
Animals from lineages G145R-15 and G145R-18 were monitored to have biochemical changes for over eighteen months without evidence of hepatocellular and renal injury, although there were histopathological changes in the liver.
In the study of vaccinated children whose mothers were positive for HBeAg, about 15% of them became anti-HBc positive. A proportion of these would be HBsAg positive andbecome the HBV carriers. About half of those were infected with variants of MHR, the most consistently observed to be G145R. Three population-based studies from Singapore, UK and USA had remarkably similar results. Mathematical modeling indicated it would be the dominant viral strain, although it could take a long time to emerge. Vaccination at birth is an ideal situation for selection of escape variants, being similar to liver transplantation, where high titer anti-HBs preparations are administered to prevent graft infection. Furthermore, naturally occurring G145R strains were reported.
To study the biological properties and pathologic conditions associated with this mutant, HBV transgenic mice whose hepatocytes expressed and replicated the virus and whose genome harbor HBV mutant G145R were produced with a terminally redundant, 1.2 genome length transgene that starts in the middle of the X gene and ends just downstream of the unique HBV polyadenylation site.
Only two of the forty-three founders were serum HBsAg-positive, and the expression level of the others was very low, which might be due to the different integrated sites of HBV DNA. In general, the exogenous gene would be integrated into multi-sites in transgenic mice. This is similar with other reports. Immunohistochemical assays of several tissues from transgenic mice revealed that HBV gene expressions were tissue-dependent, and the genes were only expressed in the liver and kidney, this is similar to HBV-infection in nature. The regulation of expression of a foreign gene in transgenic mice was subjected to different parameters, such as site of integration, influence of the mouse flanking sequences, cellular factors and structure of the integrated sequences. However, the general finding was that transgenes kept their developmental and tissue specific control of expression. Mouse genomic sequences might potentially have an influence on the level of transcription of tissue specificity. The important influence of cellular factors on expression was apparent since many hepatocytes, all the Kupffer cells, endothelial cells and bile duct epithelium in the tissue were antigen negative, although most cells in the liver probably contained integrated HBV DNA. It may be germane that hepatocyte nuclear factor 1 alpha (HNF 1 a) and retinoic acid are known to be preferentially expressed in hepatocytes. Interestingly, the positive cells were clustered like clones of cells. This was described in infected humans, and it was proposed that the synthesis of viral antigens depended on the metabolic state of cells.
HBcAg was detected both in the nuclei and in the cytoplasms of hepatocytes. The presence of HBcAg in the nuclei of hepatocytes was found in most of HBV-infected humans. Cytoplasmic location of HBcAg was also observed in some cases with active viral replication. Capsid antigens of ground squirrel hepatitis virus, duck hepatitis B virus and woodduck hepatitis virus were found almost exclusively in the cytoplasm. This may be the consequence of the constantly high level of replication of these viruses. Pre-core RNA is only used as a mRNA for the production of HBeAg that is secreted into the serum. This polypeptide can be aggregated into unstable particles but its location is in cytoplasm. Therefore, it is likely that it is responsible for the cytoplasm structure.
HBV transgenic mice showed replication of HBV in hepatocytes and had evidence of pathological lesions in the liver, which are different from other reports. Although there were some pathological characteristics of mild chronic hepatitis B in the transgenic mice, the change was obviously different from that of human chronic hepatitis B. Cellular pathological changes, such as swelling of hepatocytes and focal hepatocellular necrosis and parenchymal lymphomononuclear cell infiltrate were the main lesions. While the histological changes, such as the change or destruction of structure of hepatic lobules caused by hyperplasia of fibers, were not observed in all transgenic mice. Even fibrocytes and fibrous tissue were difficult to be observed in the portal tract, it was likely due to the difference of species. In normal mice, the borderline of hepatic lobules is not clear as that of human being’s.
Serum anti-HBs, anti-HBc and anti-HBe detected by ELISA were negative in all of the transgenic mice, suggesting that these mice were tolerant to HBsAg, HBcAg and HBeAg. This result is consistent with that of another report.
What caused the pathological changes of the liver We supposed the immune response of transgenic mice caused the pathological lesions. Some of transgenic mice were partially immunological tolerant to viral antigens. In our and another study, HBsAb could be induced by injection of HB vaccine into muscles of the HBV transgenic mice. This indicated that the immunological tolerance was not complete. We assume that although the gene of virus was integrated in the genome of mice, the time of expression of exogenous gene in thymus was a little later than the formation of immunological tolerance. Some children whose mothers were HBeAg positive did also have pathological changes in their liver, but without HBsAb in the serum.
In conclusion, HBV genome introduced by microinjection can be integrated into the mouse genome, and HBV genes can be expressed, replicated and packaged. Therefore, these lineage mutant HBV transgenic mice may be used as animal models for the study on mutant HBV and its associated biomedical issues. It can also be applied in the selection of anti-mutant HBV drugs and vaccine.
Co-correspondents: Le-Zhi Zhang and Yi-Ping Hu
Edited by Wang XL and Ren SY Proofread by Xu FM
| 1. | Merican I, Guan R, Amarapuka D, Alexander MJ, Chutaputti A, Chien RN, Hasnian SS, Leung N, Lesmana L, Phiet PH. Chronic hepatitis B virus infection in Asian countries. J Gastroenterol Hepatol. 2000;15:1356-1361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 345] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 2. | Birrer RB, Birrer D, Klavins JV. Hepatocellular carcinoma and hepatitis virus. Ann Clin Lab Sci. 2003;33:39-54. [PubMed] |
| 3. | Rabe C, Cheng B, Caselmann WH. Molecular mechanisms of hepatitis B virus-associated liver cancer. Dig Dis. 2001;19:279-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 4. | Arbuthnot P, Kew M. Hepatitis B virus and hepatocellular carcinoma. Int J Exp Pathol. 2001;82:77-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 206] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 5. | Bréchot C, Gozuacik D, Murakami Y, Paterlini-Bréchot P. Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). Semin Cancer Biol. 2000;10:211-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 226] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 6. | Lee KM, Kim YS, Ko YY, Yoo BM, Lee KJ, Kim JH, Hahm KB, Cho SW. Emergence of vaccine-induced escape mutant of hepatitis B virus with multiple surface gene mutations in a Korean child. J Korean Med Sci. 2001;16:359-362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 7. | Kidd-Ljunggren K, Miyakawa Y, Kidd AH. Genetic variability in hepatitis B viruses. J Gen Virol. 2002;83:1267-1280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 217] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 8. | Hu YP, Hu WJ, Zheng WC, Li JX, Dai DS, Wang XM, Zhang SZ, Yu HY, Sun W, Hao GR. Establishment of transgenic mouse harboring hepatitis B virus (adr subtype) genomes. World J Gastroenterol. 2001;7:111-114. [PubMed] |
| 9. | Singh M, Kumar V. Transgenic mouse models of hepatitis B virus-associated hepatocellular carcinoma. Rev Med Virol. 2003;13:243-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 10. | Kimura K, Kakimi K, Wieland S, Guidotti LG, Chisari FV. Activated intrahepatic antigen-presenting cells inhibit hepatitis B virus replication in the liver of transgenic mice. J Immunol. 2002;169:5188-5195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 88] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 11. | Sitia G, Isogawa M, Kakimi K, Wieland SF, Chisari FV, Guidotti LG. Depletion of neutrophils blocks the recruitment of antigen-nonspecific cells into the liver without affecting the antiviral activity of hepatitis B virus-specific cytotoxic T lymphocytes. Proc Natl Acad Sci U S A. 2002;99:13717-13722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 90] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Baron JL, Gardiner L, Nishimura S, Shinkai K, Locksley R, Ganem D. Activation of a nonclassical NKT cell subset in a transgenic mouse model of hepatitis B virus infection. Immunity. 2002;16:583-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 180] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 13. | Cooreman MP, Leroux-Roels G, Paulij WP. Vaccine- and hepatitis B immune globulin-induced escape mutations of hepatitis B virus surface antigen. J Biomed Sci. 2001;8:237-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 145] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 14. | Fischer L, Kalinina T, Rogiers X, Will H, Sterneck M. GLY145ARG mutation emerging under HBIG treatment in patients with recurrent HBV after liver transplantation strongly reduces viral secretion. Transplant Proc. 2001;33:3633-3636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 15. | Rodriguez-Frias F, Buti M, Jardi R, Vargas V, Quer J, Cotrina M, Martell M, Esteban R, Guardia J. Genetic alterations in the S gene of hepatitis B virus in patients with acute hepatitis B, chronic hepatitis B and hepatitis B liver cirrhosis before and after liver transplantation. Liver. 1999;19:177-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 16. | Grottola A, Buttafoco P, Del Buono MG, Cremonini C, Colantoni A, Gelmini R, Morelli C, Masetti M, Jovine E, Fruet F. Pretransplantation pre-S2 and S protein heterogeneity predisposes to hepatitis B virus recurrence after liver transplantation. Liver Transpl. 2002;8:443-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 17. | Oon CJ, Chen WN, Goo KS, Goh KT. Intra-familial evidence of horizontal transmission of hepatitis B virus surface antigen mutant G145R. J Infect. 2000;41:260-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | Chakravarty R, Neogi M, Roychowdhury S, Panda CK. Presence of hepatitis B surface antigen mutant G145R DNA in the peripheral blood leukocytes of the family members of an asymptomatic carrier and evidence of its horizontal transmission. Virus Res. 2002;90:133-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 43] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Wang Y, Wu MC, Sham JS, Tai LS, Fang Y, Wu WQ, Xie D, Guan XY. Different expression of hepatitis B surface antigen between hepatocellular carcinoma and its surrounding liver tissue, studied using a tissue microarray. J Pathol. 2002;197:610-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 20. | Jin YM, Yun C, Park C, Wang HJ, Cho H. Expression of hepatitis B virus X protein is closely correlated with the high periportal inflammatory activity of liver diseases. J Viral Hepat. 2001;8:322-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 51] [Article Influence: 2.0] [Reference Citation Analysis (0)] |