Published online Sep 15, 2004. doi: 10.3748/wjg.v10.i18.2643
Revised: February 11, 2004
Accepted: February 24, 2004
Published online: September 15, 2004
AIM: To investigate the effect of histone acetylation on regulation of p21WAF1 gene expression in human colon cancer cell lines.
METHODS: Two cell lines, Colo-320 and SW1116 were treated with either trichostatin or sodium butyrate. Expressions of p21WAF1 mRNA and protein were detected by real-time RT-PCR and Western blotting, respectively. Acetylation of two regions of p21WAF1 gene-associated histones and total cellular histones were examined by chromatin immunoprecipitation assay and Western blotting.
RESULTS: Trichostatin or sodium butyrate re-activated p21WAF1 transcription resulted in up-regulated p21WAF1 protein level in colon cancer cell lines. Those effects were accompanied by an accumulation of acetylated histones in total cellular chromatin and p21WAF1 gene-associated region of chromatin.
CONCLUSION: Histone acetylation regulates p21WAF1 expression in human colon cancer cell lines, Colo-320 and SW1116.
-
Citation: Chen YX, Fang JY, Zhu HY, Lu R, Cheng ZH, Qiu DK. Histone acetylation regulates
p21WAF1 expression in human colon cancer cell lines. World J Gastroenterol 2004; 10(18): 2643-2646 - URL: https://www.wjgnet.com/1007-9327/full/v10/i18/2643.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i18.2643
Cell cycle progression is controlled by various cyclin-dependent kinases (CDKs), whose activation is carefully regulated at multiple levels including the induction and degradation of cyclin protein, CDKs phosphorylation by cyclin-activating kinase, and the induction of CDK inhibitors (CDKIs)[1]. CDKI p21WAF1 was first cloned and characterized as an important effector that acts to inhibit CDK activity in p53 mediated cell cycle arrest in response to various agents[2]. Increased expression of p21WAF1 may play a crucial role in the G1/S phase arrest induced in transformed cells, and may prevent the progression of neoplasia[3].
Histone acetylation is emerging as a major regulatory mechanism thought to modulate gene expression by altering the accessibility of transcription factors to DNA and recent studies suggest that these alterations may also be important in the process of neoplasia formation[4]. The level of histone acetylation depends on the activity of histone acetyltransferases (HATs) and histone deacetylases (HDACs). An important approach that has been used to study the function of chromatin acetylation is the use of specific inhibitors of HDAC. Trichostatin A[5,6] (TSA, a hybrid polar compound of specific inhibitor) and sodium butyrate[4] (a short chain fatty acid produced in human colon by bacterial fermentation of carbohydrate) were reported to inhibit HDAC activity.
Previously it was revealed that acetylation of gene-associated histone or total cellular histone alone regulated p21WAF1 expression in colon cancer cell lines[7,8]. We have shown[9] that TSA or sodium butyrate induced G1 phase cell cycle arrest was linked to increased expression of p21WAF1. However, little is known about the regulation of acetylation of both gene-associated histone and total cellular histone on p21WAF1 expression in human colon cancer. It is as yet not clear about the effect of histone acetylation on p21WAF1 protein in Colo-320 and SW1116 cell lines. Therefore, in the present study, we further investigated whether TSA and sodium butyrate induced overexpression of p21WAF1 resulted from hyperacetylation of gene-associated histones and histones in total cellular chromatin in two human colon cancer cell lines, Colo-320 and SW1116.
Human colon cancer-derived cell lines Colo-320 and SW1116 were obtained from Shanghai Institute of Biochemistry and Cell Biology, SIBS, China and Shanghai Second Medical University Ruijin Hospital, respectively. Colo-320 and SW1116 cells were maintained in RPMI 1640 supplemented with 100 mL/L heat-inactivated fetal bovine serum, 2 mmol/L L-glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C in a 50 mL/L CO2 incubator.
Colon cancer cell lines were exposed to 1 μmol/L TSA or 5 mmol/L sodium butyrate (Sigma, St. Louis, MO) alone for 24 h, as described by Siavoshian et al[10]. The control cultures were treated simultaneously with phosphate-buffered saline (PBS) or alcohol (control for TSA treatment, because TSA can only be dissolved in alcohol).
Colo-320 cells were cultured as described below with or without treatment. Cells were recovered by centrifugation, washed twice with ice-cold PBS, and resuspended for lysis in 1 mL buffer A (10 mmol/L HEPES, pH7.4, 10 mmol/L KCl, 1.5 mmol/L MgCl2, 0.5 mmol/L DTT, 0.2 mmol/L PMSF, 1 μg/mL protease inhibitors, 0.25 g/L NP40) for 15 min with rotation at 4 °C and the nuclear pellet was resuspended in 100 μL buffer B (20 mmol/L HEPES, pH7.4, 420 mmol/L NaCl, 1.5 mmol/L MgCl2, 0.2 mmol/L EDTA, 250 mL/L glycerol, 0.5 mmol/L DTT, 0.2 mmol/L PMSF, 1 µg/mL protease inhibitors) for 30 min, then the soluble nuclear protein was collected by centrifugation. Fifty µg (for acetylated histone H3 and H4) or 150 µg (for p21WAF1 protein) of nuclear extracts was boiled in loading buffer (125 mmol/L Tris-HCl, pH6.8, 40 g/L SDS, 200 g/L glycerol, 0.05 g/L bromphenol blue) for 5 min and then loaded onto a 150 g/L SDS-polyacrylamide gel. After electrophoresis, proteins were transferred onto nitrocellulose membrane (0.45 µm). The following antibodies were used: rabbit polyclonal antibody against acetylated histone H3 or H4 (Upstate Biotechnology, Lake Placid, NY) and goat polyclonal antibody against p21WAF1 (C19, Santa Cruz, California). The bindings of antibodies were detected using ECL-system (Amersham Pharmacia Biotech, Piscataway, NJ, USA) and membranes were then exposed to Kodak BioMax film for 1 min. Antibody against β-actin (Sigma) in Western blot was used as a control for protein concentration.
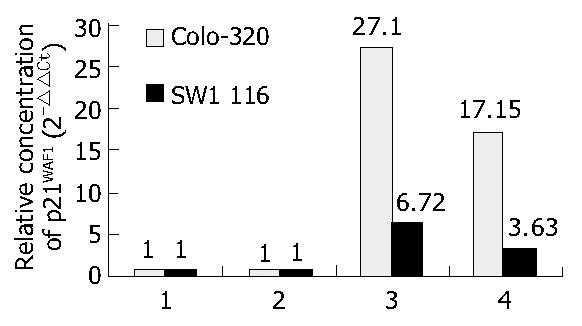
mRNA level of p21WAF1 was measured using a real-time quantitative PCR system. Total RNA samples from SW1116 and Colo-320 cells with or without treatment were prepared by TriZol Reagent. Gene-specific TaqMan probes and PCR primers were designed using Primer Express software (PE Biosystems, Foster City, CA). The sequence for forward and reverse primers and the probe are shown in Table 1. Triplicate PCR reactions were prepared for each cDNA sample. PCR consisted of 40 cycles of 95 °C denaturation (15 s) and 60 °C annealing/extension (60 s). Thermal cycling and fluorescent monitoring were performed using an ABI 7700 sequence analyzer (PE Biosystems). The point at which the PCR product is first detected above a fixed threshold, termed cycle threshold (Ct), was determined for each sample, and the average Ct of triplicate samples was calculated. To determine the quantity of gene-specific transcripts present in treated cells cDNA relative to untreated cells, their respective Ct values were first normalized by subtracting the Ct value obtained from the β-actin control (△Ct = Ct FAM-Ct VIC). The concentration of gene-specific mRNA in treated cells relative to untreated cells was calculated by subtracting the normalized Ct values obtained with untreated cells from those obtained with treated samples (△△Ct = △Ct of treated cells-DCt of untreated cells), and the relative concentration was determined (2-△△Ct). Altered mRNA expression was defined as 3-fold difference in the expression level in cells after the treatment relative to that before treatment[11].
| Gene | Primer (forward) (5’→3’) | Primer (reverse) (5’→3’) | Probe | GenBank accession number |
| p21WAF1 | CTGGAGACTCTC | GGATTAGGGCTT | ACGGCGGCAGAC | NM_078467 |
| AGGGTCGAA | CCTCTTGGA | CAGCATGA | ||
| β-actin | CTGGCACCCAGC | GGACAGCGA | ATCATTGCT | BC016045 |
| ACAATG | GGCCAGGAT | CCTCCTGAG |
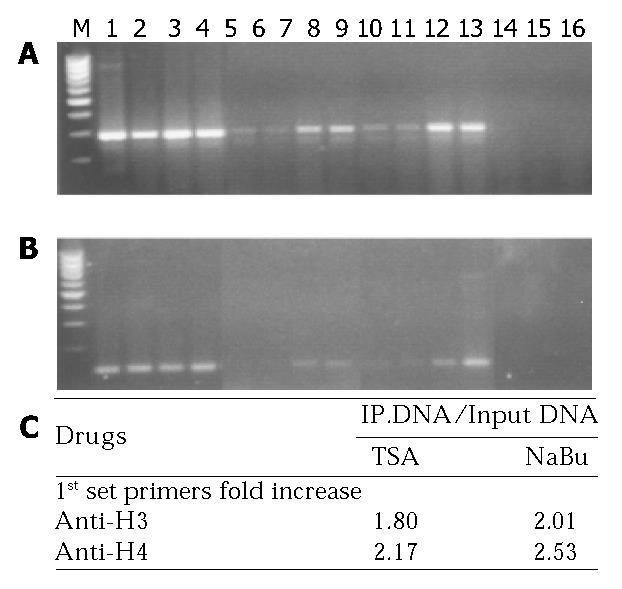
A ChIP assay kit from Upstate Biotechnology was used according to the manufacturer’s protocol and Richon’s report[12]. Colo-320 cells that were either treated with 1 µmol/L TSA or 5 mmol/L sodium butyrate for 24 h or untreated were plated at a density of 10 × 106/T25 flask. Formaldehyde was then added to the cells to a final concentration of 10 g/L, and the cells were incubated at 37 °C for 10 min. The medium was removed, and the cells were suspended in 1 mL of ice-cold PBS containing protease inhibitors [1 mmol/L phenylmethylsulfonyl fluoride (PMSF), 1 µg/mL aprotinin and 1 µg/mL pepstatin A, Boehringer Mannheim]. Cells were pelleted, resuspended in 0.2 mL of SDS lysis buffer, and incubated on ice for 10 min. Lysates were sonicated. The majority of DNAs ranged from 200 bp to 1000 bp. Debris was removed from samples by centrifugation for 10 min at 15000 g at 4 °C in a microcentrifuge. An aliquot of the chromatin preparation (200 µL) was set aside and designated as the input. Supernatants were 10-fold diluted in ChIP dilution buffer containing the protease inhibitors as above, and 80 µL of a salmon sperm DNA/protein A-agarose beads was added and incubated for 30 min at 4 °C with rocking. Beads were pelleted by centrifugation, and supernatants were placed in tubes with 10 µg of antibody against acetylated histone H3 or H4, or normal rabbit IgG, and incubated overnight at 4 °C with rotation. Salmon sperm DNA/Protein A-agarose beads (60 µL) was added, and samples were rocked for 1 h at 4 °C. Protein A complexes were centrifuged and washed 5 times for 5 min each with low salt buffer, high salt buffer, LiCl buffer and TE buffer, respectively. Immune complexes were eluted twice with 250 µL of elution buffer (10 g/L SDS/0.1 mol/L NaHCO3) for 15 min at room temperature. NaCl (5 mol/L, 20 µL) was added to the combined eluate, and the samples were incubated at 65 °C for 4 h. EDTA, Tris-HCl, pH6.5, and proteinase K were then added to the samples at a final concentration of 10 mmol/L, 40 mmol/L, and 0.04 µg/µL, respectively, and the samples were incubated at 45 °C for 1 h. Immunoprecipitated DNA (both immunoprecipitation samples and input) was recovered by phenol/chloroform extraction and ethanol precipitation and analyzed by PCR. p21WAF1-specific primers were used to carry out PCR. Sequences of two sets of primers for p21WAF1 PCR and PCR condition are shown in Table 2. The first set primer was used to amplify -576 to -293 and the second set primer was used to amplify -51 to +77 of p21WAF1 promoter and exon 1, which contained the transcription factor E2A binding sites.
| Primers | Sense (5’→3’) | Antisense (5’→3’) | Size of product and PCR condition | GenBank accession number |
| γ-actin | GGACCTGGC | GTGGCCATCTCC | 153 bp 95 °C 5 min 95 °C 1 min, | |
| TGGCCGGGACC | TGCTCGAA | 56 °C 1 min, 72 °C 1 min, 35 cycles | ||
| p21WAF1 (P1) | CGTGGTGGTGGT | CTGTCTGCA | 296 bp 95 °C 5 min 95 °C 1 min, | U24170 |
| GAGCTAGA | CCTTCGCTCCT | 58 °C 1 min, 72 °C 1 min, 35 cycles | ||
| p21WAF1 (P2) | GGTTGTATA | CTCTCACCTCCT | 128 bp 95 °C 5 min 95 °C 1 min, | U24170 |
| TCAGGGCCG | CTGAGTGC | 58 °C 1 min, 72 °C 1 min, 35 cycles |
Western blotting showed that before incubation with TSA or sodium butyrate, the levels of acetylated H3 and H4 in colo-320 cells were low. Incubation with HDAC inhibitors resulted in the accumulation of acetylated histones H3 and H4 (Figure 1).
To understand the change of p21WAF1 expression level following HDAC inhibitors treatment, we examined accumulation of mRNA and protein by RT-PCR and Western blotting. As shown in Figure 1 and Figure 2, p21WAF1 mRNA and protein were activated after treatment of TSA and sodium butyrate. In addition, Colo-320 cells had an initial increase in p21WAF1 expression to a higher level than that in SW1116 cells.
To determine whether histone acetylation reflected p21WAF1 transcription and the functional interaction between p21WAF1 and TSA or sodium butyrate treatment, ChIPs-PCR was performed. As shown in Figure 3, the densities of bands of p21WAF1 gene-associated acetylated histones H4 and H3 were higher in chromatin extracted from Colo-320 cells treated with either TSA or sodium butyrate than that from cells mock treated, either the first or the second set PCR primer.
Taken together, TSA or sodium butyrate activated the transcription of p21WAF1 through acetylation of histones H4- and H3-associated p21WAF1 promoter.
Several lines of evidence suggest that histone acetylation plays a role in transcriptional regulation, probably by altering chromatin structure[13]. Acetylation of core nucleosomal histones is regulated by the opposing activities of HATs and HDACs. The latter catalyze the removal of an acetyl group from the ε-amino group of lysine side chains of histones H2A, H2B, H3 and H4, thereby reconstituting the positive charge in lysine. Transcriptionally silent chromatin is composed of nucleosomes in which the histones have low levels of acetylation of lysine residues at their amino-terminal tails[14,15]. Acetylation of histone neutralizes the positive charge in lysine residues and disrupts nucleosome structure, allowing unfolding of the associated DNA, access by transcription factors, and changes in gene expression. Chromatin fractions enriched in actively transcribed genes are also enriched in the more highly acetylated isoforms of the core histones[16]. HDAC inhibitors appear to be selective with regard to the genes whose expression is altered[17].
Total cellular histone acetylation is also involved in the regulation of gene expression. Several studies[18] indicated that the effect of HDAC inhibitors on gene transcription was associated with an increased accumulation of acetylated histones H3 and H4 in total cellular chromatin. However, Lee’s group[19] showed an accumulation of acetylated histones H3 and H4 in total cellular chromatin after treatment with HDAC inhibitor (MS-275), but no change in the level of histone acetylation in chromatin-associated TGF-β I receptor gene. Therefore, we wanted to know whether HDAC inhibitor affected the acetylation level of histones in both gene-associated and total cellular chromatin. The data from ChIP and Western blotting suggested that p21WAF1 transcription was dependent upon acetylation at the level of chromatin, since the level of p21WAF1 promoter amplified from acetylated histone H3- or H4-associated chromatin was greater in chromatin isolated from HDAC inhibitor-treated cells than that from untreated cells. Accumulation of acetylated p21WAF1-assocaited histones induced by HDAC inhibitors was higher than that in total cellular chromatin, although there was accumulation of acetylated histones H3 and H4 in total cellular chromatin.
It is noteworthy that, the level of acetylated histones H3 and H4 at the domain containing the transcriptional start site in p21WAF1 promoter and the binding sites of E2A was significantly higher than that at another domain or total cellular chromatin analyzed. The result of our observations suggested that p21WAF1 expression could be activated by histone acetylation of its transcription start domain in promoter. Therefore, a possible mechanism involved the binding of transcription factor E2A to p21WAF1 promoter at transcription start site of acetylated p21WAF1 gene-associated histones H3 and H4, and the enhancement of p21WAF1 gene transcription. It is known that histone acetylation can be targeted to specific promoters by gene-specific activator. E2A transcription factor belongs to the basic helix-loop-helix family of proteins[20], which contains a conserved basic region responsible for DNA binding and a helix-loop-helix domain for dimerization[21]. E2A binds p21WAF1 at the domain nearby TATA box in the promoter. TATA box-independent transcription of the p21WAF1 promoter has been previously reported[22]. The proximal p21WAF1 promoter contains a TATA box[23]. Some reports indicated that p21WAF1 was up-regulated by E2A binding to HTLV-1-infected T cells[20]. Moreover, overexpression of E2A proteins, such as E47 has been shown to induce p21WAF1 promoter activity independent of p53 binding sites[24,25].
Also, we showed that the levels of p21WAF1 mRNA and protein in colon cancer cells were very low, even difficult to detect before treatment. In Colo-320, p21WAF1 mRNA was increased by 27.1-fold and 17.15-fold after 1 μmol/L TSA and 5 mmol/L sodium butyrate treatment, respectively. Accordingly, the protein level of p21WAF1 was elevated. Similar effects were shown in SW1116 cells (data not shown). Our data about TSA or sodium butyrate inducing p21WAF1 mRNA and protein expression are consistent with previous reports[12].
In summary, this study demonstrated that HDAC inhibitor, TSA or sodium butyrate, activated the expressions of p21WAF1 mRNA and protein, and this increased expression was associated with an accumulation of acetylated histones in total cellular chromatin and the chromatin of p21WAF1 gene in these two colon cancer cell lines. It has been shown that p21WAF1 expression is reduced in adenomas and colorectal carcinomas. Our observations support the claim for the therapeutic potential of HDAC inhibitors in the treatment of colorectal carcinoma, because there is probably no mutation of the p21WAF1 gene in colorectal cancer.
Thanks are given to Mr. En-Lin Li, Ms. Wei-Qi Gu and Ms. Hong-Yin Zhu for performing cell culture and the real-time PCR assay and Western blotting.
| 1. | Sherr CJ. Mammalian G1 cyclins and cell cycle progression. Proc Assoc Am Physicians. 1995;107:181-186. [PubMed] |
| 2. | Dulić V, Kaufmann WK, Wilson SJ, Tlsty TD, Lees E, Harper JW, Elledge SJ, Reed SI. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1030] [Cited by in RCA: 1070] [Article Influence: 33.4] [Reference Citation Analysis (6)] |
| 3. | Kim JS, Lee S, Lee T, Lee YW, Trepel JB. Transcriptional activation of p21(WAF1/CIP1) by apicidin, a novel histone deacetylase inhibitor. Biochem Biophys Res Commun. 2001;281:866-871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Archer SY, Hodin RA. Histone acetylation and cancer. Curr Opin Genet Dev. 1999;9:171-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 194] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 5. | Magdinier F, Wolffe AP. Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc Natl Acad Sci USA. 2001;98:4990-4995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 156] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 6. | Xiao H, Hasegawa T, Isobe K. p300 collaborates with Sp1 and Sp3 in p21(waf1/cip1) promoter activation induced by histone deacetylase inhibitor. J Biol Chem. 2000;275:1371-1376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 145] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 7. | Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. 2002;132:1012-1017. [PubMed] |
| 8. | Hinnebusch BF, Henderson JW, Siddique A, Malo MS, Zhang W, Abedrapo MA, Hodin RA. Transcriptional activation of the enterocyte differentiation marker intestinal alkaline phosphatase is associated with changes in the acetylation state of histone H3 at a specific site within its promoter region in vitro. J Gastrointest Surg. 2003;7:237-244; discussion 244-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 9. | Chen YX, Fang JY, Lu J. Epigenetics is involved in regulation of cell cycle and expression of tumor suppressor genes in human colon cancer cells. Chin J Dig Dis. 2003;4:105-110. |
| 10. | Siavoshian S, Segain JP, Kornprobst M, Bonnet C, Cherbut C, Galmiche JP, Blottière HM. Butyrate and trichostatin A effects on the proliferation/differentiation of human intestinal epithelial cells: induction of cyclin D3 and p21 expression. Gut. 2000;46:507-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 218] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 11. | Scanlan MJ, Welt S, Gordon CM, Chen YT, Gure AO, Stockert E, Jungbluth AA, Ritter G, Jäger D, Jäger E. Cancer-related serological recognition of human colon cancer: identification of potential diagnostic and immunotherapeutic targets. Cancer Res. 2002;62:4041-4047. [PubMed] |
| 12. | Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci USA. 2000;97:10014-10019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 878] [Cited by in RCA: 887] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 13. | Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2143] [Cited by in RCA: 2180] [Article Influence: 75.2] [Reference Citation Analysis (0)] |
| 14. | Wolffe AP, Pruss D. Targeting chromatin disruption: Transcription regulators that acetylate histones. Cell. 1996;84:817-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 285] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 15. | Vettese-Dadey M, Grant PA, Hebbes TR, Crane- Robinson C, Allis CD, Workman JL. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508-2518. [PubMed] |
| 16. | Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1345] [Cited by in RCA: 1415] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 17. | Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 1996;5:245-253. [PubMed] |
| 18. | Mishra N, Reilly CM, Brown DR, Ruiz P, Gilkeson GS. Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse. J Clin Invest. 2003;111:539-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 296] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 19. | Lee BI, Park SH, Kim JW, Sausville EA, Kim HT, Nakanishi O, Trepel JB, Kim SJ. MS-275, a histone deacetylase inhibitor, selectively induces transforming growth factor beta type II receptor expression in human breast cancer cells. Cancer Res. 2001;61:931-934. [PubMed] |
| 20. | de La Fuente C, Deng L, Santiago F, Arce L, Wang L, Kashanchi F. Gene expression array of HTLV type 1-infected T cells: Up-regulation of transcription factors and cell cycle genes. AIDS Res Hum Retroviruses. 2000;16:1695-1700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 56] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 21. | Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1812] [Cited by in RCA: 2072] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 22. | Datto MB, Yu Y, Wang XF. Functional analysis of the transforming growth factor beta responsive elements in the WAF1/Cip1/p21 promoter. J Biol Chem. 1995;270:28623-28628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 355] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 23. | Owen GI, Richer JK, Tung L, Takimoto G, Horwitz KB. Progesterone regulates transcription of the p21(WAF1) cyclin- dependent kinase inhibitor gene through Sp1 and CBP/p300. J Biol Chem. 1998;273:10696-10701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 272] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 24. | Prabhu S, Ignatova A, Park ST, Sun XH. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol. 1997;17:5888-5896. [PubMed] |
| 25. | Fang JY, Lu YY. Effects of histone acetylation and DNA methylation on p21( WAF1) regulation. World J Gastroenterol. 2002;8:400-405. [PubMed] |
Edited by Chen WW and Zhu LH Proofread by Xu FM