Published online Sep 15, 2004. doi: 10.3748/wjg.v10.i18.2632
Revised: February 18, 2004
Accepted: February 21, 2004
Published online: September 15, 2004
AIM: Mitotic cell death has been focused on in tumor therapy. However, the precise mechanisms underlying it remain unclear. We have reported previously that enediyne antibiotic lidamycin induces mitotic cell death at low concentrations in human epithelial tumor cells. The aim of this study was to investigate the possible link between centrosome dynamics and lidamycin-induced mitotic cell death in human hepatoma BEL-7402 cells.
METHODS: Growth curve was established by MTT assay. Cell multinucleation was detected by staining with Hoechst 33342. Flow cytometry was used to analyze cell cycle. Aberrant centrosomes were detected by indirect immunofluorescence. Western blot and senescence-associated β-galactosidase (SA-β-gal) staining were used to analyze protein expression and senescence-like phenotype, respectively.
RESULTS: Exposure of BEL-7402 cells to a low concentration of lidamycin resulted in an increase in cells containing multiple centrosomes in association with the appearance of mitotic cell death and activation of SA-β-gal in some cells, accompanied by the changes of protein expression for the regulation of proliferation and apoptosis. The mitochondrial signaling pathway, one of the major apoptotic pathways, was not activated during mitotic cell death. The aberrant centrosomes contributed to the multipolar mitotic spindles formation, which might lead to an unbalanced division of chromosomes and mitotic cell death characterized by the manifestation of multi- or micronucleated giant cells. Cell cycle analysis revealed that the lidamycin treatment provoked the retardation at G2/M phase, which might be involved in the centrosome overduplication.
CONCLUSION: Mitotic cell death and senescence can be induced by treatment of BEL-7402 cells with a low concentration of lidamycin. Centrosome dysregulation may play a critical role in mitotic failure and ultimate cell death following exposure to intermediate dose of lidamycin.
- Citation: Liang YX, Zhang W, Li DD, Liu HT, Gao P, Sun YN, Shao RG. Mitotic cell death in BEL-7402 cells induced by enediyne antibiotic lidamycin is associated with centrosome overduplication. World J Gastroenterol 2004; 10(18): 2632-2636
- URL: https://www.wjgnet.com/1007-9327/full/v10/i18/2632.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i18.2632
Liver cancer is one of the most malignant tumors in the world[1,2]. Surgical resection is considered the most effective but not the most popular method for the treatment of hepatocellular carcinoma (HCC). Chemotherapy is indicated for a large member of HCC patients. Mitotic cell death is a cell death form different from apoptosis, on which has been focused in tumor therapy. It is also known as mitotic catastrophe or delayed reproductive death, and can be activated by radiation or antitumor agents at low doses or concentrations[3-5]. Mitotic cell death is frequently characterized by enlargement of cell volume, appearance of multi- or micronucleation, and arrest in G2/M phase of cell cycle. Finally, these cells underwent death. Thus far, little is known about the mechanism responsible for mitotic cell death. Some researchers considered that defects in mitotic machinery, such as multiple rounds of DNA synthesis without an intervening cytokinesis, and chromosome missegregation, might play a key role in the process of lethal nuclear fragmentation[6]. Previous reports have suggested that the absence or delay of the G1/S checkpoint and the subsequent absence of interphase apoptosis coupled to this checkpoint contribute to mitotic cell death[7,8].
The centrosome, representing the major microtubule organizing centre in eukaryotic cells, contains a pair of centrioles surrounded by pericentriolar material. The centrosome duplicates once during each cell cycle. To complete the normal cell cycle, the centrosome duplication cycle and the centrosome quantity must be precisely regulated to couple the other events of cell cycle[9]. If centrosome replication deviates from cycles of DNA synthesis and mitotic division, an unsuccessful mitosis will come out with the features associated with the formation of aberrant centrosomes and multiple mitotic spindles, and unbalanced chromosome segregation[10].
Enediyne antibiotics have been focused on their potent antitumor activity due to their unique ability to damage the DNA of tumor cells by inducing single strand (SSB) and/or double strand (DSB) breaks through free radical attacks on the deoxyribose moieties in DNA[11]. Lidamycin (also designated as C1027) is a member of the enediyne antibiotic family, which was isolated from a Streptomyces globisporus C1027 strain in China[12,13]. Lidamycin consists of a chromophore and an apoprotein, and the former has the ability to attack DNA, whereas the latter plays the role as a protecting protein[14]. The biological effects induced by lidamycin and ionizing radiation are similar[11]. Previous reports have shown that lidamycin is highly cytotoxic toward tumor cells[14-16]. As an attempt to investigate the mechanisms of lidamycin-induced mitotic cell death in human hepatoma BEL-7402 cells, we treated cells with lidamycin at low concentrations, and discovered centrosome overduplication, multipolar mitotic spindle formation, multinucleation, delayed reproductive death and changed patterns of protein expression associated with the regulation of proliferation and apoptosis. These results indicate that mitotic cell death in BEL-7402 cells induced by lidamycin is associated with centrosome overduplication independently of mitochondria pathway.
Lidamycin was generously provided by Professor Lian-Fang Jin from our institute, and stored at -20 °C as a 100 μmol/L stock solution in 9 g/L NaCl solution.
Human hepatoma BEL-7402 cells (obtained from the Key Laboratory of Cell Proliferation and Regulation Biology of the Ministry of Education, Beijing Normal University) were cultured in DMEM (Gibcol BRL) supplemented with 100 mL/L fetal bovine serum (HyClone), 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C in the presence of 50 mL/L CO2.
Growth curves establishments were performed at a 5-d interval as previously described[17] with some modifications. Totally 2.0 × 103 cells were seeded into 96-well plates and then treated with lidamycin for 2 h. A 12 μL MTT (5 mg/mL) was added to each well before assay and incubated for an additional 4 h at 37 °C, followed by treatment with 100 μL of 0.01 mol/L HCl-100 g/L SDS overnight. The value at each time point was read on a Microplate Reader (Model 550, Bio-Rad) at λ570nm.
Subconfluent cells were continuously incubated for 72 h after exposure to lidamycin for 2 h, and then were stained by the DNA-specific fluorescent dye Hoechst 33342 (2 μg/mL) (Sigma) for 15 min at 37 °C. Next, cells were washed once, kept in PBS, and observed using a fluorescence microscope (BH2 system, Olympus) equipped with a λ455nm filter.
Cells were exposed to 0.5 nmol/L lidamycin for 2 h and then incubated in fresh, drug-free medium. Following a 3-d incubation, cells including the floating and the attached were harvested and washed with cold PBS twice. Cell suspensions were fixed in 700 mL/L ethanol at 4 °C overnight. Next, the fixed cells were washed twice in PBS and incubated with 50 μg/mL RNase (Sigma) for 30 min at 37 °C. Samples were then stained with 50 μg/mL propidium iodide (Sigma) in the dark at 4 °C for 30 min, and analyzed on a fluorescence-activated cell sorter (EPICS XL, Coulter).
Cells were grown on coverslips. After 3 d following 2-h lidamycin treatment, the cells were washed in PHEM buffer (60 mmol/L PIPES, 25 mmol/L HEPES, 10 mmol/L EGTA and 2 mmol/L MgCl2) twice briefly, and incubated with a permeabilization buffer (5 mL/L Triton X-100 in PHEM buffer) for 90 s. Then, the cells were fixed in 37 g/L paraform in PHEM buffer for 15 min at room temperature. After washed in PBS 3 times, the cells were incubated with a blocking solution (50 g/L defatted dry milk and 0.5 mL/L Tween-20 in PBS) for 30 min and used for indirect immunofluorescence. The primary antibodies included anti-α-tubulin monoclonal antibody (Zymed) and anti-centrin polyclonal antibody[18] (kindly provided by Professor Da-Cheng He, the Key Laboratory of Cell Proliferation and Regulation Biology of the Ministry of Education, Beijing Normal University). Rhodamine-labeled goat anti-mouse antibody (Zymed) and Fluoresceinisothiocyanate-labeled goat anti-rabbit antibody (Zymed) were used as second antibodies. The microscope slides were mounted with glycerol mounting medium (900 mL/L glycerol and 100 mL/L PBS) and observed under a laser-scanning microscope (IX-70 system, Olympus). The cell with three or more centrosomes was considered aberrant.
Cells were treated with 0.1 nmol/L or 0.5 nmol/L lidamycin for 2 h and continuously maintained for 72 h. The attached cells were fixed in 5 mL/L glutaraldehyde and stained for SA-β-gal activity using X-gal at pH6.0 as previously described[19].
Cells incubated with lidamycin at 37 °C for 2 h and then allowed to recover for 72 h at 37 °C were harvested and washed in PBS. The cells were lysed on ice in lysis buffer (100 mmol/L Tris, pH6.8, 25 g/L SDS, 100 mL/L β-mercaptoethanol, 1 mmol/L phenylmethylsulfonyl fluoride, and 100 mL/L glycerol) for 10 min, followed by ultrasonication. The cell lysates were cleared by centrifugation, and the protein concentration was estimated using the Bradford method with bovine serum albumin as a standard. Western blot analysis was performed as a protocol described previously[20]. In brief, equal amounts of protein were electrophoresed on SDS-polyacrylamide gel (Fluka) and transferred onto a nitrocellulose membrane (Hybond-P, Amersham Pharmacia) for blotting with primary antibodies including anti-Bax (N-20, Santa Cruz), anti-Smac (a kind gift from Dr. Xiao-Dong Wang, University of Texas Southwestern Medical Center, Dallas, USA), anti-cyclin B1 (GNS-1, Santa Cruz), anti-p16 (16P04, NeoMarkers), anti-Rb (C-15, Santa Cruz), anti-p53 (DO-1, Santa Cruz), anti-p21 (F-5, Santa Cruz), and anti-actin (I-19, Santa Cruz) antibodies. Secondary antibodies conjugated with horseradish peroxidase (Amersham Pharmacia). Enhanced chemiluminescence (ECL Western Blot Kit, Amersham Pharmacia) was used according to the manufacturer’s instructions.
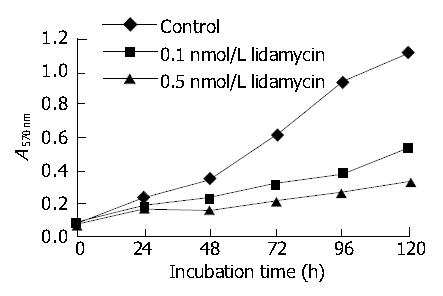
Inhibition of cell growth and proliferation was measured by the MTT test. Exposing BEL-7402 cells to 0.1 and 0.5 nmol/L lidamycin resulted in a dose-dependent inhibition of cell growth (Figure 1).
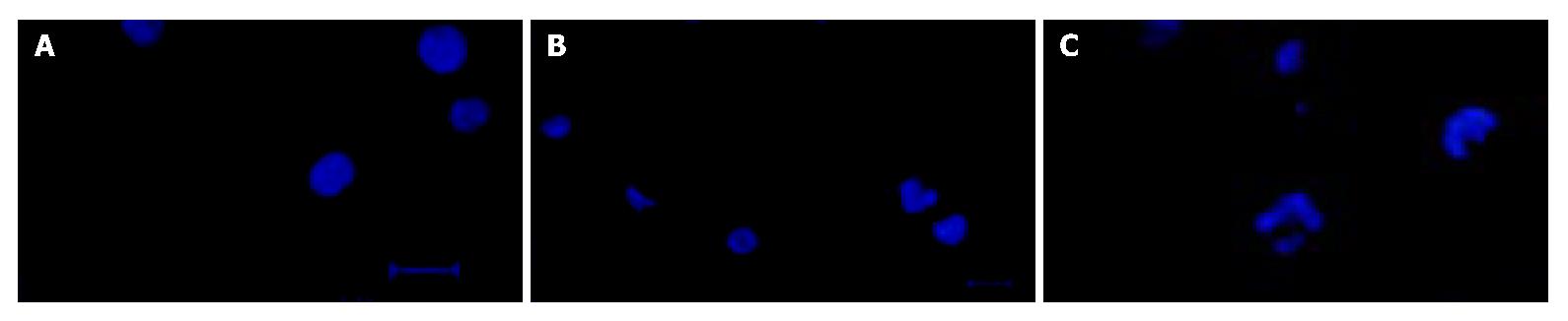
With 0.1 nmol/L lidamycin for 2 h, followed by a 72-h incubation in drug-free medium, the treated BEL-7402 cells displayed a unique and atypical chromatin condensation characterized by appearance of small “dots” representing segregated condensed chromatin without apoptotic bodies (Figure 2B). Moreover, we did not observe detachment of these cells from the monolayer during the process of chromatin condensation. After 72-h incubation, multinucleation (three or more nuclei), one of the main features of mitotic cell death, occurred at 0.5 nmol/L lidamycin-treated BEL-7402 cells (Figure 2C).
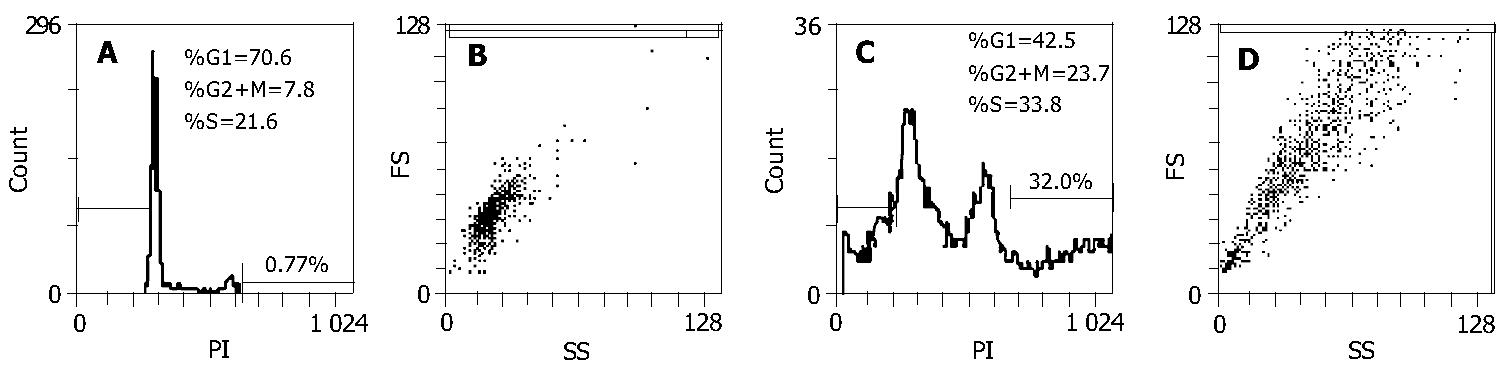
The biochemical and cytological changes of multinucleated giant cells remain poorly understood. To further characterize the etiology of mitotic cell death, we analyzed the cell cycle and DNA content of the lidamycin-treated BEL-7402 cells by flow cytometry. The cells were exposed to a low concentration of lidamycin for 2 h. At 72 h after treatment, ~23.7% of BEL-7402 cells arrested in G2/M phase, and the cells with > 4N DNA content were detected at 32.0% (Figure 3C).
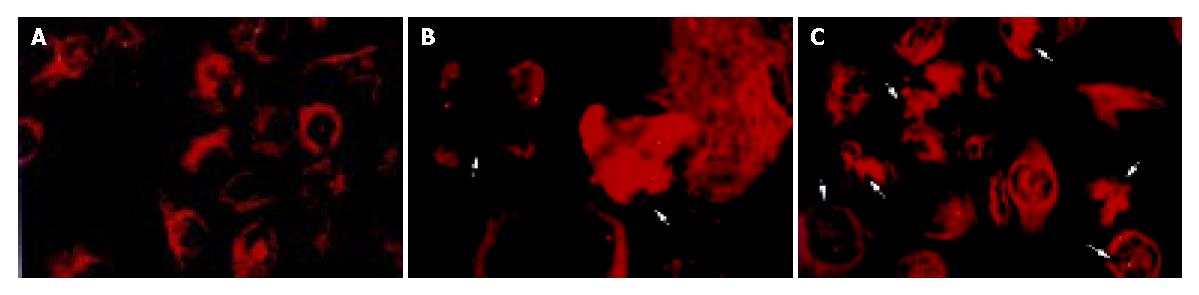
Immunofluorescence microscopy revealed that the abnormalities of multiple centrosomes and multipolar mitotic spindles markedly increased in BEL-7402 cells induced by lidamycin at a low concentration. Co-staining with antibodies to centrin and α-tubulin indicated that the increased centrosomes were localized at each pole of the multiple spindles, and the cells not deriving from equal division appeared (Figure 4B, Figure 4C). However, to untreated cells, these defects rarely displayed (Figure 4A).
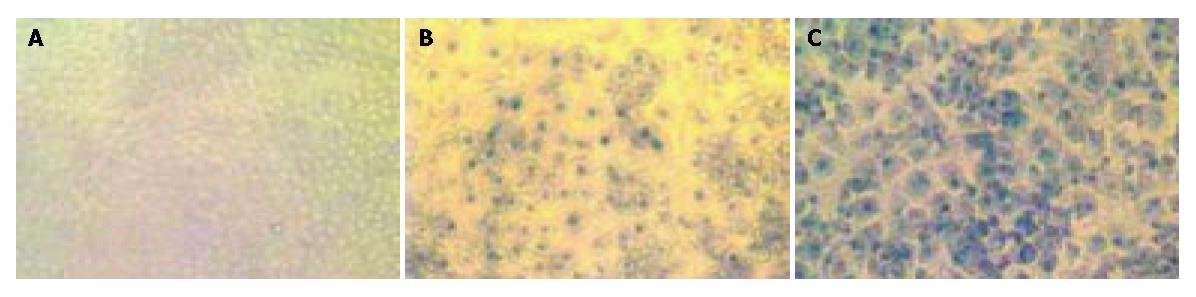
The induction of SA-β-gal activity and mitotic cell death were thought to be independent events[21]. To investigate the effects of mitotic cell death on senescence-like phenotype of the lidamycin-treated cells, we observed SA-β-gal expression, a senescence marker, at 72 h after lidamycin treatment. The treated cells showed phenotypic changes that resembled features of normal senescence, including enlarged and flattened morphology, increased granularity, vacuolization, and enhanced SA-β-gal- positive cells (Figure 5). Moreover, the induction of senescence-like phenotype in BEL-7402 cells was increased dose-dependently.
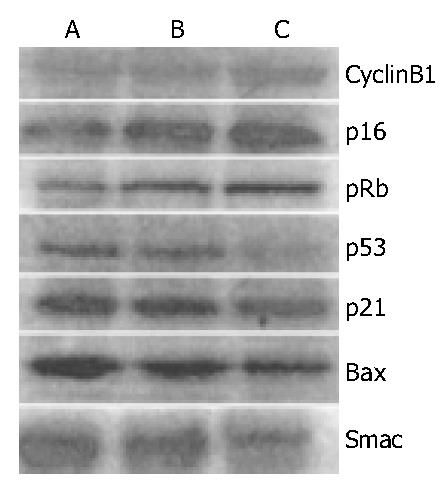
To understand some molecular changes that led to mitotic cell death, cell cycle proteins were analysed by Western blot analysis in lidamycin-treated cells. In BEL-7402 cells, lidamycin induced an increase in the levels of cyclin B1, p16 and pRb, meanwhile a decrease in expression of p53 and p21 (Figure 6).
To confirm that mitotic cell death induced by lidamycin was distinguished from typical apoptosis, and detect the correlation between them, proteins associated with apoptosis were examined. Bax is a regulator of apoptosis[20]. Smac, an inhibitor of caspase suppressors, promotes cytochrome c-induced activation of caspases by sequestering the inhibitor of apoptosis protein family[22]. As is shown in Figure 6, the protein levels of Bax and Smac declined in lidamycin-treated BEL-7402 cells. We were unable to detect the proteolytically activated caspase-3 and caspase-9, and found no significant alterations of the blots representing caspase-3 precursor and caspase-9 precursor after exposure to low concentrations of lidamycin in BEL-7402 cells (data not shown).
HCC is one of the most common malignant neoplasms in the world[1]. The incidence of HCC in China exceeds 100000 per year, and at least 110000 HCC-related deaths occur every year. Lidamycin is a highly potent cytotoxic antitumor agent. We treated human hepatoma BEL-7402 cells with lidamycin at concentrations in the nanomolar range, and assessed the potential role of centrosome in lidamycin-induced mitotic cell death. The results indicated a series of abnormal events including centrosome overduplication, formation of multipolar mitotic spindles, multinucleation, and eventual mitotic catastrophe. In this study, we have described for the first time the association between centrosomes and enediyne antibiotics-induced cell death in human hepatoma cells.
Thus far, the modes of cell death induced by lidamycin can be divided into two classes, one is apoptosis, and the other is mitotic cell death[5,23]. It has been reported that lidamycin can act directly as an endonuclease without dependence on caspase activities and is considered as an apoptosis-mimetic agent, at high concentrations[24]. Cells exposed to low concentrations of lidamycin lost reproductive integrity due to inappropriate entry into mitosis, and apparently exhibited the morphological and biochemical changes associated with mitotic cell death: enlarged cell shape, multi- or micronucleation, accumulation of karyotypic abnormalities, and a G2/M arrest[5]. Apoptosis, mitotic cell death and irreversible cell cycle arrest may all contribute to cell death after lidamycin treatment. The exact mechanism of mitotic cell death is unclear, and only a few studies have attempted to elucidate the effects of lidamycin on cells at moderate concentrations[5,11]. In the present study, we used low-dose lidamycin to treat BEL-7402 cells, and mitotic cell death was observed predominantly after treatment. We demonstrate that lidamycin can induce multiple centrosomes which may be responsible for the assembly of multipolar mitotic spindles and the chromosomes missegregation. Most of the cells containing multiple nuclear fragments were temporarily viable but reproductively dead. However, in some cases, the cells undergoing mitotic death initiated endocycles, restituted mitosis and finally survival[8]. We plan to continue this study to confirm the link between lidamycin-induced mitotic cell death and centrosome overduplication by using centrosome inhibitors and vectors containing antisense mRNAs to centrosome related proteins.
Because centrosome duplication was closely associated with DNA replication, cytokinesis and cell cycle regulation[25,26], we analyzed the cell cycle progression of lidamycin-treated BEL-7402 cells to detect relationship between centrosome dysregulation and cell cycle distribution as well as to confirm the appearance of multinucleated cells, which is one of the main features of mitotic cell death. We found that lidamycin induced centrosome overduplication associated with induction of G2/M arrest. Previous reports showed that centrosome replication could dissociate from DNA synthesis cycle and mitotic division[27], and cell cycle block in G2/M phase might be related to abnormal centrosome accumulation[10], and endocycles starting from G2/M arrest could produce endopolyploid cells[7]. We suppose that lidamycin-induced DNA replication cycle retardation in BEL-7402 cells could be helpful to trigger centrosome overduplication.
Analysis of gene expression may provide further insights into the molecular mechanisms mediating mitotic cell death. Cyclin B1, a component of the mitosis-promoting factor, plays an important role in G2/M regulation by forming a complex with p34cdk1 to phosphorylate various substrates necessary for mitosis. The cells with the morphological features of mitotic catastrophe frequently undergo up-regulation of cyclin B1 level[28,29], which is consistent with our results. Bax expression is a regulator of apoptosis. Ordinarily action of Bax facilitates apoptosis[30]. However, our present study demonstrated that the levels of Bax and Smac both decreased along with no proteolytic activation of caspase-3 and caspase-9, and no DNA ladder was obtained (data not shown) after lidamycin treatments, which suggested that the mitochondrial apoptosis pathway might not be activated. The undetectable typical apoptosis is not caused by Bcl-2 involvement in BEL-7402 cells exposed to low concentrations of lidamycin (data not shown) and some other genes might play an essential role in this response. p21 is a p53-regulated protein. p53 inhibition was shown to increase mitotic death[31]. We noticed decreased levels of p53 and p21 in BEL-7402 cells after lidamycin treatment. However, the expression of p16 and pRb proteins was upregulated in BEL-7402 cells, which might explain the increased intensity of staining for SA-β-gal in lidamycin-treated cells, since p16 is closely related to induction of senescence-like phenotype[32]. Based on data presented here, we propose that the G2/M arrest of BEL-7402 cells may not be mediated by a classically driven cell cycle checkpoint mechanism correlated with p53. From a therapeutic standpoint, centrosome dysregulation might provide a valuable anticancer target. Further study to identify signaling pathways to mitotic cell death in tumor cells and normal somatocytes would help to improve the efficacy of HCC therapy with a low systemic toxicity.
We thank Drs. Lian-Fang Jin, Da-Cheng He and Xiao-Dong Wang for providing lidamycin and antibodies to centrin and Smac. We thank Wei-Li Cai for technical assistance. Hong-Bin Deng, Jian-Ming Jiang and Guo Li are acknowledged for their helpful discussions.
| 1. | Bosch FX, Ribes J, Borràs J. Epidemiology of primary liver cancer. Semin Liver Dis. 1999;19:271-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 671] [Cited by in RCA: 672] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 2. | Shi YJ, Gong JP, Liu CA, Li XH, Mei Y, Mi C, Huo YY. Construction of a targeting adenoviral vector carrying AFP promoter for expressing EGFP gene in AFP-producing hepatocarcinoma cell. World J Gastroenterol. 2004;10:186-189. [PubMed] |
| 3. | Jonathan EC, Bernhard EJ, McKenna WG. How does radiation kill cells. Curr Opin Chem Biol. 1999;3:77-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 210] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 4. | Ianzini F, Mackey MA. Spontaneous premature chromosome condensation and mitotic catastrophe following irradiation of HeLa S3 cells. Int J Radiat Biol. 1997;72:409-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 66] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 5. | He QY, Liang YY, Wang DS, Li DD. Characteristics of mitotic cell death induced by enediyne antibiotic lidamycin in human epithelial tumor cells. Int J Oncol. 2002;20:261-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 6. | Allen CE, Wu LC. Downregulation of KRC induces proliferation, anchorage independence, and mitotic cell death in HeLa cells. Exp Cell Res. 2000;260:346-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 7. | Erenpreisa J, Cragg MS. Mitotic death: a mechanism of survival A review. Cancer Cell Int. 2001;1:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 93] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 8. | Grafi G. Cell cycle regulation of DNA replication: the endoreduplication perspective. Exp Cell Res. 1998;244:372-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 30] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 9. | Winey M. Keeping the centrosome cycle on track. Genome stability. Curr Biol. 1996;6:962-964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 10. | Shono M, Sato N, Mizumoto K, Minamishima YA, Nakamura M, Maehara N, Urashima T, Saimura M, Qian L, Nishio S. Effect of serum depletion on centrosome overduplication and death of human pancreatic cancer cells after exposure to radiation. Cancer Lett. 2001;170:81-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 11. | Dziegielewski J, Beerman TA. Cellular responses to the DNA strand-scission enediyne C-1027 can be independent of ATM, ATR, and DNA-PK kinases. J Biol Chem. 2002;277:20549-20554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 12. | Hu JL, Xue YC, Xie MY, Zhang R, Otani T, Minami Y, Yamada Y, Marunaka T. A new macromolecular antitumor antibiotic, C-1027. I. Discovery, taxonomy of producing organism, fermentation and biological activity. J Antibiot (Tokyo). 1988;41:1575-1579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 93] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 13. | Otani T, Minami Y, Marunaka T, Zhang R, Xie MY. A new macromolecular antitumor antibiotic, C-1027. II. Isolation and physico-chemical properties. J Antibiot (Tokyo). 1988;41:1580-1585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 56] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | Shao RG, Zhen YS. [Relationship between the molecular composition of C1027, a new macromolecular antibiotic with enediyne chromophore, and its antitumor activity]. Yaoxue Xuebao. 1995;30:336-342. [PubMed] |
| 15. | Zhen YS, Ming XY, Yu B, Otani T, Saito H, Yamada Y. A new macromolecular antitumor antibiotic, C-1027. III. Antitumor activity. J Antibiot (Tokyo). 1989;42:1294-1298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 84] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 16. | Xu YJ, Zhen YS, Goldberg IH. C1027 chromophore, a potent new enediyne antitumor antibiotic, induces sequence-specific double-strand DNA cleavage. Biochemistry. 1994;33:5947-5954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 65] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 17. | Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38285] [Cited by in RCA: 40049] [Article Influence: 931.4] [Reference Citation Analysis (1)] |
| 18. | Chen XB, Wang YC, Li YZ, Cui JT. Identification of a novel resi-dent protein in centrosome. Kexue Tongbao. 2000;45:1302-1307. |
| 19. | Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363-9367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5281] [Cited by in RCA: 5889] [Article Influence: 190.0] [Reference Citation Analysis (0)] |
| 20. | Liang Y, Yan C, Schor NF. Apoptosis in the absence of caspase 3. Oncogene. 2001;20:6570-6578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 177] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 21. | Chang BD, Broude EV, Dokmanovic M, Zhu H, Ruth A, Xuan Y, Kandel ES, Lausch E, Christov K, Roninson IB. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 1999;59:3761-3767. [PubMed] |
| 22. | Lademann U, Cain K, Gyrd-Hansen M, Brown D, Peters D, Jäättelä M. Diarylurea compounds inhibit caspase activation by preventing the formation of the active 700-kilodalton apoptosome complex. Mol Cell Biol. 2003;23:7829-7837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 23. | Jiang B, Li DD, Zhen YS. Induction of apoptosis by enediyne antitumor antibiotic C1027 in HL-60 human promyelocytic leukemia cells. Biochem Biophys Res Commun. 1995;208:238-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 24. | Wang Z, He Q, Liang Y, Wang D, Li YY, Li D. Non-caspase-mediated apoptosis contributes to the potent cytotoxicity of the enediyne antibiotic lidamycin toward human tumor cells. Biochem Pharmacol. 2003;65:1767-1775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 25. | Hinchcliffe EH, Cassels GO, Rieder CL, Sluder G. The coordination of centrosome reproduction with nuclear events of the cell cycle in the sea urchin zygote. J Cell Biol. 1998;140:1417-1426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 72] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 26. | Lacey KR, Jackson PK, Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc Natl Acad Sci U S A. 1999;96:2817-2822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 306] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 27. | Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol. 1995;130:105-115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 248] [Cited by in RCA: 249] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 28. | Nabha SM, Mohammad RM, Dandashi MH, Coupaye-Gerard B, Aboukameel A, Pettit GR, Al-Katib AM. Combretastatin-A4 prodrug induces mitotic catastrophe in chronic lymphocytic leukemia cell line independent of caspase activation and poly(ADP-ribose) polymerase cleavage. Clin Cancer Res. 2002;8:2735-2741. [PubMed] |
| 29. | Hyun JW, Cheon GJ, Kim HS, Lee YS, Choi EY, Yoon BH, Kim JS, Chung MH. Radiation sensitivity depends on OGG1 activity status in human leukemia cell lines. Free Radic Biol Med. 2002;32:212-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 30. | Okuno S, Shimizu S, Ito T, Nomura M, Hamada E, Tsujimoto Y, Matsuda H. Bcl-2 prevents caspase-independent cell death. J Biol Chem. 1998;273:34272-34277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 127] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 31. | Chang BD, Xuan Y, Broude EV, Zhu H, Schott B, Fang J, Roninson IB. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene. 1999;18:4808-4818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 288] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 32. | Drayton S, Rowe J, Jones R, Vatcheva R, Cuthbert-Heavens D, Marshall J, Fried M, Peters G. Tumor suppressor p16INK4a determines sensitivity of human cells to transformation by cooperating cellular oncogenes. Cancer Cell. 2003;4:301-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 74] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
Edited by Zhang JZ Proofread by Zhu LH and Xu FM