Published online Aug 1, 2004. doi: 10.3748/wjg.v10.i15.2223
Revised: December 23, 2003
Accepted: January 8, 2004
Published online: August 1, 2004
AIM: To evaluate the inhibition effect of HCV NS5A on p53 transactivation on p21 promoter and explore its possible mechanism for influencing p53 function.
METHODS: p53 function of transactivation on p21 promoter was studied with a luciferase reporter system in which the luciferase gene is driven by p21 promoter, and the p53-DNA binding ability was observed with the use of electrophoretic mobility-shift assay (EMSA). Lipofectin mediated p53 or HCV NS5A expression vectors were used to transfect hepatoma cell lines to observe whether HCV NS5A could abrogate the binding ability of p53 to its specific DNA sequence and p53 transactivation on p21 promoter. Western blot experiment was used for detection of HCV NS5A and p53 proteins expression.
RESULTS: Relative luciferase activity driven by p21 promoter increased significantly in the presence of endogenous p53 protein. Compared to the control group, exogenous p53 protein also stimulated p21 promoter driven luciferase gene expression in a dose-dependent way. HCV NS5A protein gradually inhibited both endogenous and exogenous p53 transactivation on p21 promoter with increase of the dose of HCV NS5A expression plasmid. By the experiment of EMSA, we could find p53 binding to its specific DNA sequence and, when co-transfected with increased dose of HCV NS5A expression vector, the p53 binding affinity to its DNA gradually decreased and finally disappeared. Between the Huh 7 cells transfected with p53 expression vector alone or co-transfected with HCV NS5A expression vector, there was no difference in the p53 protein expression.
CONCLUSION: HCV NS5A inhibits p53 transactivation on p21 promoter through abrogating p53 binding affinity to its specific DNA sequence. It does not affect p53 protein expression.
- Citation: Gong GZ, Jiang YF, He Y, Lai LY, Zhu YH, Su XS. HCV NS5A abrogates p53 protein function by interfering with p53-DNA binding. World J Gastroenterol 2004; 10(15): 2223-2227
- URL: https://www.wjgnet.com/1007-9327/full/v10/i15/2223.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i15.2223
Hepatitis C virus (HCV) is recognized as a major causative agent leading to chronic hepatitis and cirrhosis[1-3], which have a close relationship with the development of hepatocellular carcinoma (HCC)[4,5]. HCV is a positive single-stranded RNA virus belonging to the Flaviridae family, and its genome only contains a single long open reading frame encoding a large polyprotein precursor that is thereafter processed by a combination of cellular and viral proteases into several mature proteins, including three or four structural proteins (core, E1, E2/P7) and at least 6 nonstructural proteins (NS): NS2, NS3, NS4A, NS4B, NS5A, NA5B[6]. HCV NS5A as a nonstructural protein does not assemble into the HCV particles, but still has a lot of functions in the HCV replication and the development of chronic liver disease and hepatocellular carcinoma. Recent studies showed that HCV NS5A could interact with a number of cellular proteins including PKR, p53, Grb2 and Cdk1[7-10], and enhance expression of some genes related to cell proliferation such as PCNA, NF-κ B, Stat-3, SRCAP and IL-8[11-14], indicating that HCV NS5A has a critical role in promoting cell proliferation and malignant transformation. HCV NS5A prevents p53 and TNF-a mediated apoptosis[15-17], and possibly perturbs the DNA repair when cells are treated with DNA damage agents including viruses, toxins or physical damage. HCV NS5A is important for the HCV replication. It can form a complex with an SNARE-like protein, hVAP-33 and HCV NS5B[18] and can associate with ER and Golgi complex, and the amphipathic helix (AH) of the HCV NS5A is necessary for this membrane localization and for HCV RNA replication[19,20]. Interaction of HCV NS5A with La protein can also maintain and benefit HCV RNA replication and HCV protein translation[21,22]. Sequence mutation in HCV NS5A might be a reason for the responsiveness in the patient who received IFN treatment[23-25]. The patients with wild type of interferon sensitive-determining region (ISDR) in HCV NS5A usually have a lower responsive rate to the IFN therapy, and the mechanism is that the wild type ISDR can bind PKR, which is induced by IFN and has an anti-viral activity, and can disrupt PKR function[7,26,27]. The tumor suppressor p53 protein has been reported to possess a number of important functions. Activated p53 can transactivate a lot of target genes, maintain normal cell cycle through inducing apoptosis and repairing damaged DNA, and suppressing oncogenic transformation. In this study we explored the interaction between HCV NS5A and p53 and the mechanism by which the HCV NS5A abrogated the p53 transactivation.
PC53-NS3, Pwwp-luc and Pwwp-mut-luc were kind gifts from Professor Vogelstein (The Johns Hopkins University). PC53-NS3 is an eukaryotic expression vector that is constructed by cloning normal human p53 cDNA into pCMV plasmid. Pwwp-luc carries luciferase reporter gene driven by p21 promoter that contains the specific consensus DNA sequence binding to p53 protein. Pwwp-mut-luc is approximately the same vector as Pwwp -luc except for deletion mutation introduced in the p53 binding sequence of p21 promoter. PCNS5A is made in Dr. Siddiqui’s laboratory (University of Colorado, USA) through inserting of HCV NS5A cDNA into PCMV plasmid[12]. The liver carcinoma cell lines Huh7 with p53 gene mutation and HepG2 with wild-type p53 gene were from ATCC(USA) and maintained in Dulbecco’s Modified Eagle Medium (Life Technologies, USA) complemented with 100 mL/L fetal calf serum. The cells were transfected by the individual plasmids with Lipofectin reagent (Life Technologies, USA) when they became 60-70% confluent. Forty-eight hours after transfection, the cells were harvested for the analysis of relative luciferase activity and electrophoretic mobility shift assay (EMSA).
After 48 h of transfection, cells in the plates were washed two times with PBS, and then 750 μL lysis buffer (0.1 mmol/L K2HPO4, pH 7.8, 10 mmol/L DTT, 5 g/L NP-40) was added onto plates. The plates were then placed on ice for 15 min and then cells were transferred into a clean 1.5 -mL Eppendorf tube. After centrifugation at 15 000 g for 10 min, the supernatants were collected for further experiment. For relative luciferase activity assay, 0.4 mL determination buffer (0.1 mmol/L K2HPO4, pH 7.8, 0.15 mmol/L MgSO4, 10 mmol/L DTT, 0.5 mmol/L ATP) and the different volumes of supernatant containing equivalent protein were put into 5- mL glass tube to observe the times of luminescence within 10 s using a luminometer (Optocomp II, MGM,USA). The experiment was repeated three times and each in triplicate.
In this experiment, we investigated the ability of p53 binding to the specific consensus DNA sequence, and studied if the binding could be influenced by HCV NS5A. Briefly, this experiment was done as following: (1) Labeling of DNA probe: 50 pmol chemically synthesized oligonucleotide 5’ - GGACATGCCCGG GCATGTCC -3’ which is the specific consensus DNA sequence to bind p53, 2 µL γ -P32 ATP (74 000 GBq/mmoL), 20 U T4 polynucleotide kinase, 2 µL reaction buffer and incubated for 30 min at 37 °C, then 60 µL dH2O was added to the mixture and the labeled oligonucleotides were purified from the free γ -P32 ATP by use of Sephadex-G25. The purified P32-labeled DNA probe was stored at -20 °C until use. (2) Nuclear protein isolation: After transfection with different plasmids for 48 h, cells were washed 3 times with PBS and scrapped down from the plates and then transferred into 1.5-mL Eppendorf tubes and washed 3 times again with PBS. Lysis buffer 30 µL (20 mmol/L Hepes, pH 7.9, 10 mmol/L KCl, 10 mmol/L Na3VO4, 1 mmol/L ETDA, 100 mL/L glycerol, 1 mmol/L PMSF, 3 g/L aprotinin, 1 g/L leupeptin, 1 mmol/L DTT, 2 g/L NP-40, 20 mmol/L NaF) was added to suspend the cells. After 15 min on ice, and centrifugation at 15 000 × g for 1 min, the supernatant was discarded and the pellet resuspended in 20 µL nuclear extract buffer (20 mmol/L Hepes, pH7.9, 10 mmol/L KCl, 420 mmol/L NaCl, 10 mmol/L Na3VO4, 1 mmol/L ETDA, 200 mL/L glycerol, 1 mmol/L PMSF, 3 g/L aprotinin, 1 g/L leupeptin, 1 mmol/L DTT, 20 mmol/L NaF). After incubation at 4 °C for 30 min. with mild agitation, and centrifugation at 15 000 g for 5 min, aliquot supernatants were collected and stored at -70 °C until use. (3) Assessment of p53 binding to DNA probe: For each reaction tube, the followings were added: 2 µg nuclear extraction protein, 2 µL labeled probe containing p53 binding sequence, 1.5 µL EMSA reaction buffer (50 mmol/L NaCl, 0.5 mmol/L ETDA, 40 mL/L glycerol, 1 mmol/L MgCl2, 1 mmol/L DTT, 10 mmol/L Tris-Cl, pH 7.5, 0.05 g/L poly (dI -dC) ) and dH2O to a total volume of 15 µL. After incubation for 30 min on ice, 20 µL of samples was loaded onto60 g/L undenatured PAGE and run at 160 V for approximately 6 h. The gel was dried and autoradiographed at -70 °C for about 16 h.
Cell lysates of Huh7 cells transfected with PC53-NS3 or co-transfected with PCNS5A were made by routine method. The same amount of total protein from the cell lysate was used to load onto 120 g/L PAGE, and then transferred onto PV (polyvinylidene difluoride) membrane. After blocked in block solution (20 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl, 3 g/L PVP, 5 g/L Tween 20) for 2 h, the membrane was probed by anti-HCV NS5A or anti-p53, washed and incubated with the second enzyme-labeled antibody, and then observed by using an ECL kit (Boegringer Mannhein Co., Germany).
Statistical significance of differences between the 2 groups was determined by applying Student’s t test or two-sample t test. Statistical significance of differences in more than 3 groups was determined by applying one-way ANOVA. A P value less than 0.05 was considered significant.
Relative luciferase activities in Pwwp-luc-transfected HepG2 cells ( 3.95 × 105) were significantly higher than that in Pwwp-mut-luc group (0.60 × 105, t = 5.92, P < 0.01). Compared to Pwwp-luc-transfected HepG2 cells, the relative luciferase activities in the HepG2 cells co-transfected with Pwwp -luc and PCNS5A decreased in a PCNS5A dose- dependent manner (F = 20.71, P < 0.01), suggesting that HCV NS5A protein can inhibit the transactivation of endogenous p53 on p21 promoter. In contrast, the relative luciferase activities of the HepG2 cells co-transfected with Pwwp-mut-luc and PCNS5A had little change (F = 0.76, P > 0.05) in comparison with Pwwp-mut-luc-transfected HepG2 cells. Those results indicated that endogenous p53 has a transactivation on p21 promoter, and HCV NS5A down-regulated p21 promoter activity through inhibiting endogenous p53 activity (Table 1).
| Group | Relative luciferase activity | F | P |
| pwwp-luc (1 µg) | |||
| + pCNS5A (0 µg) | 3.49 × 105b | 20.71 | < 0.01 |
| + pCNS5A (0.5 µg) | 2.69 × 105 | ||
| + pCNS5A (1.0 µg) | 1.62 × 105 | ||
| + pCNS5A (2.0 µg) | 0.58 × 105 | ||
| pwwp-mut-luc (1.0 μg) | |||
| + pCNS5A (0 µg) | 0.60 × 105 | 0.76 | > 0.05 |
| + pCNS5A (0.5 µg) | 0.65 × 105 | ||
| + pCNS5A (1.0 µg) | 0.62 × 105 | ||
| + pCNS5A (2.0 µg) | 0.64 × 105 |
The mean of relative luciferase activity in the Huh7 cells individually transfected with Pwwp-luc and Pwwp-mut-lu was 0.47 × 105 and 0.45 × 105 respectively and there were no statistically differences between these two groups (t = 1.16, P > 0.05). This might be because p53 gene in Huh7 cells has been mutated and p53 protein has lost its normal transactivation function, therefore, no matter what happened to p21 promoter (wild type or mutated), the expression of p21 promoter driven luciferase gene was not affected. When Pwwp-luc or Pwwp-mut-luc was co- transfected with PC53-NS3 into Huh7 cells, respectively, the relative luciferase activities in the Pwwp-luc + PC53-NS3 group greatly increased (χ = 5.63 × 105), but the relative luciferase activities in the Pwwp-mut-luc + PC53-NS3 group only mild increased (χ = 0.50 × 105), and a remarkable variance between the two groups existed (t = 10.12, P < 0.01). These results indicated that exogenous p53 should have a strong transactivation on p21 promoter. In order to see if HCV NS5A protein has any effect on the exogenous p53 transactivation, we used PCNS5A, P C53-NS3and Pwwp-luc to co-transfect Huh7 cells, and the result showed that the relative luciferase activities in the PCNS5A+PC53-NS3+Pwwp-luc group cells decreased in a PCNS5A dose-dependent way (F = 24.23, P < 0.01). PCNS5A and Pwwp-luc were also used to transfect Huh7 cells to find out if HCV NS5A had a direct effect on p21 promoter. The result showed that HCV NS5A did not increase or decrease the relative luciferase activity. From those experiments we proposed that HCV NS5A down-regulated p21 promoter activity by inhibiting exogenous p53 activity, and not by HCV NS5A’ s direct effect on p21 promoter (Table 2 and Table 3).
| Group | Relative luciferase activity | F | P |
| pwwp-luc (1 µg) | 0.47 × 105ab | 24.23 | < 0.01 |
| +pC53-NS3 (1 µg) | 5.63 × 105b | ||
| +pCNS5A (0.5 µg) | 1.61 × 105 | ||
| +pCNS5A (1.0 µg) | 0.69 × 105 | ||
| pwwp-mut-luc (1 µg) | 0.45 × 105a | 0.69 | > 0.05 |
| +pC53-NS3 (1 µg) | 0.50 × 105 | ||
| +pCNS5A (0.5 µg) | 0.43 × 105 | ||
| +pCNS5A (1.0 µg) | 0.48 × 105 |
| Group | Relative luciferase activity | F | P |
| pwwp-luc (1.0 µg) | 0.44 × 105 | 0.52 | > 0.05 |
| + pCNS5A (0.5 µg) | 0.46 × 105 | ||
| + pCNS5A (1.0 µg) | 0.43 × 105 | ||
| pwwp-luc (1.0 µg) | 0.56 × 105 | 28.16 | < 0.01 |
| + pC53-NS (0.5 µg) | 2.68 × 105 | ||
| + pC53-NS (1.0 µg) | 4.84 × 105 |
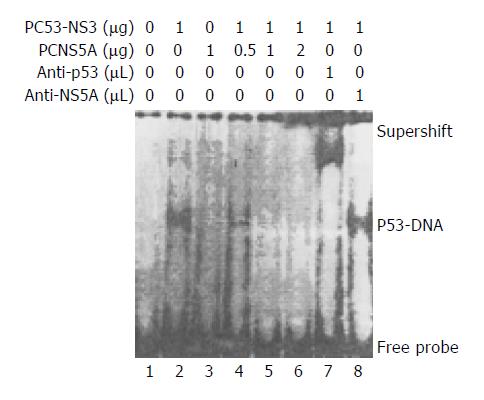
Through EMSA, the band of specific p53 and DNA probe complex was found in the nuclear lysates prepared from Huh7 cells transfected with PC53-NS3, and verified by the appearance of the supershift after addition of the monoclonal anti-p53 (a generous gift of Dr. Harris, USA) or polyclonal anti-HCV NS5A antibody (a generous gift of Professor Rice, USA) in the EMSA mixtures. This p53-probe complex band did not occur in the nuclear lysates prepared from Huh7 cells untransfected with plasmids or only transfected with PCNS5A, suggesting there is wild-type p53 protein expressed in these cells. In the next experiment, we used different doses of PCNS5A and constant amount (1 µg) of PC53-NS3 to co-transfect Huh7 cells. The p53-probe band became gradually weakened and finally disappeared with increasing doses of PCNS5A, suggesting that HCV NS5A could inhibit p53 transactivation through disruption of the binding of p53 to its specific consensus DNA (Figure 1).
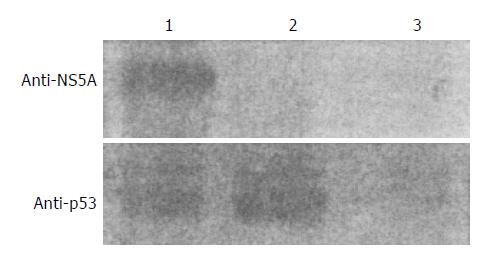
p53 protein was expressed at about the same level between the Huh7 cells transfected with p53 expression vector and the Huh7 cells co-transfected with PCNS5A and p53 expression vectors (Figure 2). This result indicates that HCV NS5A does not affect p53 protein expression.
HCV NS5A is a phosphoprotein that exists in two different forms of Mr 56 000 and 58 000 with modifications of serine residues, and Mr 58 000 form is produced by additional phosphorylation of the Mr 56 000 form[28-30]. Initial studies found that HCV NS5A contains acidic and proline-rich amino acids in its carboxyl-terminal, and this structure feature resembles eukaryotic transcription activator[31,32]. Later studies showed that the HCV NS5A with amino-terminal deleted (1-146) mutant did have a strong transcriptional activation, but the whole length HCV NS5A showed no apparent transcriptional activity[33]. The potential tansactivation of HCV NS5A has attracted great attention and become the research focus of the nonstructural proteins of HCV due to its important role in the pathogenesis of chronic liver disease and even hepatocellular carcinoma. Clinical observations suggested the responsiveness of patients to IFN treatment might be related to HCV NS5A ISDR sequence mutations[23-25]. Several experiments from Katze´s laboratory and others confirmed that HCV NS5A could repress RNA dependent protein kinase (PKR) function, by which HCV can escape the effect of IFN, and also inhibit apoptosis and promote cell proliferation[7,26,27]. When PKR is dysfunctioned, sustained expression of eIF -2α in cells results in cell growth and differentiation, and induces malignant transformation and the development of tumors in nude mice. ISDR of HCV NS5A can bind the domain of PKR and prevent the PKR dimerizing which is the critical process to activate PKR function. Mapping studies found that ISDR and the adjacent C-terminal 26 amino acids can form a heterodimer with PKR. The effect of HCV NS5A on the protection of apoptosis induced by TNF-α and p53 was further identified[16,17]. In the present study, we explored the possible effect of HCV NS5A on the transactivation of p53. Luciferase reporter gene driven by p21 promoter was used in this experiment, which showed that both endogenous and exogenous p53 protein could transactivate p21 promoter as expected. But, when HCV NS5A protein was introduced, by transfection or co-transfection with HCV NS5A expressing vector, the p21 promoter function activated by p53 was greatly depressed. This finding is in agreement with previous reports[8,11]. To further understand the mechanism of interaction between HCV NS5A and p53, we did an EMSA experiment for p53 binding to its consensus DNA probe, to see if HCV NS5A could disturb the binding reaction. The result showed that HCV NS5A disrupted p53 binding to its DNA probe, and with the increase of dosage of HCV NS5A expressing vector transfected, inhibition of the binding seemed more remarkable. All the above results suggested HCV NS5A depressed the p53 transactivation by interrupting the binding between p53 and its specific DNA. Recent studies also evidenced that HCV NS5A could physically associate with p53 and form a complex, which might be the reason why p53 binding to its DNA was disrupted[8,34]. Further, we wanted to know whether p53 protein expression was affected by the presence of HCV NS5A. Through western blot, we found that after co-transfection of HCV NS5A expressing vector, the level of p53 protein expression in the cells did not show any change compared to empty vector transfected cells. This result was also similarly found in another report[11]. In summary, according to our experiments and previous reports by others, HCV NS5A represses p53 transactivation function by disrupting p53 binding to its specific DNA through HCV NS5A and p53 complex formation, not by the disturbance of the p53 protein expression. HCV NS5A can interact with many cellular factors in the host cells such as PKR, p53, Bcl2 and disrupt their functions[7,8,26,27,34,35], which are related to the processes of apoptosis. HCV NS5A can also interact with unknown cellular factors and induce the expression of genes such as SRCAP, NF -κ B and PCNA[11-13], which are related to the promotion of cell proliferation. HCV NS5A seems to be a very active protein interacting with host cellular proteins and taking a major part in modulating cell growth and death, as well as in the development of hepatocellular carcinoma.
We would like to thank Dr. A Siddiqui and G Waris from University of Colorado School of Medicine for their help in this study.
| 1. | Farci P. Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome [Science 1989; 244: 359-362]. J Hepatol. 2002;36:582-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4996] [Cited by in RCA: 4670] [Article Influence: 126.2] [Reference Citation Analysis (1)] |
| 2. | Kuo G, Choo QL, Alter HJ, Gitnick GL, Redeker AG, Purcell RH, Miyamura T, Dienstag JL, Alter MJ, Stevens CE. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2495] [Cited by in RCA: 2345] [Article Influence: 63.4] [Reference Citation Analysis (1)] |
| 3. | Seeff LB. Natural history of hepatitis C. Hepatology. 1997;26:21S-28S. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 295] [Article Influence: 10.2] [Reference Citation Analysis (4)] |
| 4. | Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547-6549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 826] [Cited by in RCA: 840] [Article Influence: 23.3] [Reference Citation Analysis (1)] |
| 5. | Di Bisceglie AM, Lyra AC, Schwartz M, Reddy RK, Martin P, Gores G, Lok AS, Hussain KB, Gish R, Van Thiel DH. Hepatitis C-related hepatocellular carcinoma in the United States: influence of ethnic status. Am J Gastroenterol. 2003;98:2060-2063. [PubMed] |
| 6. | Clarke B. Molecular virology of hepatitis C virus. J Gen Virol. 1997;78:2397-2410. [PubMed] |
| 7. | Gale M, Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM, Korth MJ, Polyak SJ, Gretch DR, Katze MG. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208-5218. [PubMed] |
| 8. | Majumder M, Ghosh AK, Steele R, Ray R, Ray RB. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J Virol. 2001;75:1401-1407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 209] [Article Influence: 8.4] [Reference Citation Analysis (1)] |
| 9. | Tan SL, Nakao H, He Y, Vijaysri S, Neddermann P, Jacobs BL, Mayer BJ, Katze MG. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc Natl Acad Sci USA. 1999;96:5533-5538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 172] [Article Influence: 6.4] [Reference Citation Analysis (1)] |
| 10. | Arima N, Kao CY, Licht T, Padmanabhan R, Sasaguri Y, Padmanabhan R. Modulation of cell growth by the hepatitis C virus nonstructural protein NS5A. J Biol Chem. 2001;276:12675-12684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 93] [Article Influence: 3.7] [Reference Citation Analysis (1)] |
| 11. | Ghosh AK, Steele R, Meyer K, Ray R, Ray RB. Hepatitis C virus NS5A protein modulates cell cycle regulatory genes and promotes cell growth. J Gen Virol. 1999;80:1179-1183. [PubMed] |
| 12. | Gong G, Waris G, Tanveer R, Siddiqui A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa B. Proc Natl Acad Sci USA. 2001;98:9599-9604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 485] [Cited by in RCA: 502] [Article Influence: 20.1] [Reference Citation Analysis (1)] |
| 13. | Ghosh AK, Majumder M, Steele R, Yaciuk P, Chrivia J, Ray R, Ray RB. Hepatitis C virus NS5A protein modulates transcription through a novel cellular transcription factor SRCAP. J Biol Chem. 2000;275:7184-7188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 73] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 14. | Polyak SJ, Khabar KS, Paschal DM, Ezelle HJ, Duverlie G, Barber GN, Levy DE, Mukaida N, Gretch DR. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J Virol. 2001;75:6095-6106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 240] [Article Influence: 9.6] [Reference Citation Analysis (1)] |
| 15. | Lan KH, Sheu ML, Hwang SJ, Yen SH, Chen SY, Wu JC, Wang YJ, Kato N, Omata M, Chang FY. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene. 2002;21:4801-4811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 177] [Article Influence: 7.4] [Reference Citation Analysis (1)] |
| 16. | Majumder M, Ghosh AK, Steele R, Zhou XY, Phillips NJ, Ray R, Ray RB. Hepatitis C virus NS5A protein impairs TNF-mediated hepatic apoptosis, but not by an anti-FAS antibody, in transgenic mice. Virology. 2002;294:94-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 89] [Article Influence: 3.7] [Reference Citation Analysis (1)] |
| 17. | Ghosh AK, Majumder M, Steele R, Meyer K, Ray R, Ray RB. Hepatitis C virus NS5A protein protects against TNF-alpha mediated apoptotic cell death. Virus Res. 2000;67:173-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 2.6] [Reference Citation Analysis (2)] |
| 18. | Tu H, Gao L, Shi ST, Taylor DR, Yang T, Mircheff AK, Wen Y, Gorbalenya AE, Hwang SB, Lai MM. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology. 1999;263:30-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 187] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 19. | Elazar M, Cheong KH, Liu P, Greenberg HB, Rice CM, Glenn JS. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J Virol. 2003;77:6055-6061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 140] [Article Influence: 6.1] [Reference Citation Analysis (1)] |
| 20. | Brass V, Bieck E, Montserret R, Wölk B, Hellings JA, Blum HE, Penin F, Moradpour D. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J Biol Chem. 2002;277:8130-8139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 283] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 21. | Houshmand H, Bergqvist A. Interaction of hepatitis C virus NS5A with La protein revealed by T7 phage display. Biochem Biophys Res Commun. 2003;309:695-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 22. | He Y, Tan SL, Tareen SU, Vijaysri S, Langland JO, Jacobs BL, Katze MG. Regulation of mRNA translation and cellular signaling by hepatitis C virus nonstructural protein NS5A. J Virol. 2001;75:5090-5098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 2.0] [Reference Citation Analysis (1)] |
| 23. | Schiappa DA, Mittal C, Brown JA, Mika BP. Relationship of hepatitis C genotype 1 NS5A sequence mutations to early phase viral kinetics and interferon effectiveness. J Infect Dis. 2002;185:868-877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 24. | Hung CH, Lee CM, Lu SN, Lee JF, Wang JH, Tung HD, Chen TM, Hu TH, Chen WJ, Changchien CS. Mutations in the NS5A and E2-PePHD region of hepatitis C virus type 1b and correlation with the response to combination therapy with interferon and ribavirin. J Viral Hepat. 2003;10:87-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 25. | Watanabe H, Enomoto N, Nagayama K, Izumi N, Marumo F, Sato C, Watanabe M. Number and position of mutations in the interferon (IFN) sensitivity-determining region of the gene for nonstructural protein 5A correlate with IFN efficacy in hepatitis C virus genotype 1b infection. J Infect Dis. 2001;183:1195-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 50] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 26. | Tan SL, Katze MG. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology. 2001;284:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 153] [Article Influence: 6.1] [Reference Citation Analysis (1)] |
| 27. | Gale M, Kwieciszewski B, Dossett M, Nakao H, Katze MG. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J Virol. 1999;73:6506-6516. [PubMed] |
| 28. | Kaneko T, Tanji Y, Satoh S, Hijikata M, Asabe S, Kimura K, Shimotohno K. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem Biophys Res Commun. 1994;205:320-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 164] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 29. | Hirota M, Satoh S, Asabe S, Kohara M, Tsukiyama-Kohara K, Kato N, Hijikata M, Shimotohno K. Phosphorylation of nonstructural 5A protein of hepatitis C virus: HCV group-specific hyperphosphorylation. Virology. 1999;257:130-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 30. | Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980-3986. [PubMed] |
| 31. | Chung KM, Song OK, Jang SK. Hepatitis C virus nonstructural protein 5A contains potential transcriptional activator domains. Mol Cells. 1997;7:661-667. [PubMed] |
| 32. | Tanimoto A, Ide Y, Arima N, Sasaguri Y, Padmanabhan R. The amino terminal deletion mutants of hepatitis C virus nonstructural protein NS5A function as transcriptional activators in yeast. Biochem Biophys Res Commun. 1997;236:360-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 2.0] [Reference Citation Analysis (1)] |
| 33. | Kato N, Lan KH, Ono-Nita SK, Shiratori Y, Omata M. Hepatitis C virus nonstructural region 5A protein is a potent transcriptional activator. J Virol. 1997;71:8856-8859. [PubMed] |
| 34. | Qadri I, Iwahashi M, Simon F. Hepatitis C virus NS5A protein binds TBP and p53, inhibiting their DNA binding and p53 interactions with TBP and ERCC3. Biochim Biophys Acta. 2002;1592:193-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 35. | Chung YL, Sheu ML, Yen SH. Hepatitis C virus NS5A as a potential viral Bcl-2 homologue interacts with Bax and inhibits apoptosis in hepatocellular carcinoma. Int J Cancer. 2003;107:65-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 79] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
Edited by Zhu LH and Chen WW Proofread by Xu FM