Published online Aug 1, 2004. doi: 10.3748/wjg.v10.i15.2179
Revised: February 23, 2004
Accepted: March 2, 2004
Published online: August 1, 2004
AIM: To study the differential expression of proteins in normal and cancerous gastric tissues, and further identify new molecular markers for diagnosis and prognosis of gastric carcinoma, as well as develop new therapeutic targets of the disease.
METHODS: Matched pairs of tissues from 6 gastric cancer patients were analyzed for their two-dimensional electrophoresis (2DE) profiles. Soluble fraction proteins from human normal and cancerous gastric tissue were separated in the first dimension by isoelectric focusing on immobilized pH gradient (IPG, pH3-10) strips, and by 125 g/L sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) in the second dimension with silver nitrate staining. Protein differential expression was analyzed by use of image analysis software to find out candidates for gastric cancer-associated proteins.
RESULTS: Nine protein spots overexpressed in tumor tissues as compared with noncancerous regions. In the next step, 9 tumor-specific spots were cut off from Coomassie Brilliant Blue staining gels, digested in gel with L-1-tosylamide-2-phenylethyl chloromethyl ketone (TPCK)-trypsin. Protein identification was done by peptide mass fingerprinting with matrix assisted laser desorption/ ionization-time of flight-mass spectrometry (MALDI-TOF-MS). In total, 5 tumor-specific protein spots corresponding to 5 different polypeptide chains were identified, including annexin V, carbonic anhydrase, prohibitin, fibrin beta and fibrinogen fragment D. Among these 5 spots, the potential significance of the differential expressions is discussed.
CONCLUSION: Differential expression analysis of proteomes may be useful for the development of new molecular markers for diagnosis and prognosis of gastric carcinoma.
- Citation: Wang KJ, Wang RT, Zhang JZ. Identification of tumor markers using two-dimensional electrophoresis in gastric carcinoma. World J Gastroenterol 2004; 10(15): 2179-2183
- URL: https://www.wjgnet.com/1007-9327/full/v10/i15/2179.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i15.2179
Gastric cancer is a prevalent tumor worldwide. It is a multistage process involving multiple factors in aetiology and many gene-environment interactions. It is important to emphasize the heterogeneity of the histological background on which the tumor develops. Methods have been developed to identify tumor associated antigens such as molecular cloning in expression system or using a biochemical strategy based on the extraction of antigenic peptides bound to major histocompatibility complex class 1 molecules from tumor cells. These methods have allowed the recognition of certain human tumor antigens[1]. Several tumor markers of gastric cancer have been identified[2-4], including Lewis antigen, sulfomucin, CA50. However, no evidence has been obtained indicating that the detection of these markers precedes clinical diagnosis of gastric cancer. Proteomics studies of clinical tumor samples have led to the identification of cancer-specific protein markers, which provides a basis for developing new methods for early diagnosis and early detection of cancers as well as clues to understanding the molecular mechanism of cancer progression[5]. In order to identify proteins that elicit humoral responses in gastric cancer patients, proteome-based approach was used. A number of proteins from gastric tumor tissues were separated by 2DE and identified by using mass spectrometry. It includes the systematic cataloging of protein expression on a large scale. Such studies could help elucidate the molecular mechanism of cellular events associated with cancer progression, such as cellular signaling[6,7].
Six pairs of primary, and advanced poorly differentiated gastric adenocarcinoma tissues and corresponding adjacent noncancerous gastric tissues were obtained with informed consent from patients who underwent gastrectomy at the First Affiliated Hospital of Zhengzhou University and Beijing Cancer Hospital. Cancer samples were obtained from the “core” part of the tumor to avoid the adjacent noncancerous tissue. For each of the normal tissues, surface epithelium was selectively procured by dissection with special care for minimal contamination of nonepithelial cells, and samples were immediately snap-frozen in liquid nitrogen. They were classified histologically according to Lauren’s classification after H &E staining.
Fragments of normal and malignant tissues were sharp dissected and homogenized with a homogenizer in 2 mL fresh lysis buffer [2 g/L dithiothreilol (Amersham Bioscience, USA), 200 mL/L trichloroacetic acid (Sigma, USA) and 800 g/L acetone], then placed into tubes at 4-8 °C for 8-10 h. The mixture was centrifuged at 1000 r/min for 5 min to remove tissue and cell debris, then centrifuged in a Beckman TL-100 table top ultracentrifuge at 430000 g in a TLA-100.2 rotor for 30 min at 4 °C. The supernatant was taken as soluble fraction. Protein was lyophilized, resuspended in isoelectric focusing gel rehydration solution {7 mol/L urea, 2 mol/L thiourea, 40 g/L 3-([3-cholamidopropyl] dimethylammonio) - 1-propanesulfonate(CHAPS), 50 mmol/L dithiothreilol, 20 g/L IPG buffer pH 3-10, L} and stored at - 80 °C until use. These were used as the 2DE samples for the soluble fraction. Protein concentration of 2-DE samples was estimated according to a commercial Bradford reagent. BSA was used as standard.
First-dimension bioelectric focusing was carried out on Multiphor II system basically as described by the manufacturer Amersham Bioscience Inc. Samples containing up to 200 µg protein for analytical gels were diluted to up to 450 µL with dehydration solution (8 mol/L urea, 20 g/L CHAPS, 100 mol/L dithiothreilol, 5 g/L IPG buffer). Pre-cast Immobilized pH gradient (IPG) strip (24 cm, pH 3-10, linear gradient) was used for the first-dimensional separation. Strips were applied by overnight rehydration at 50 V, and for 1 h at 1 000 V. Then a gradient was applied from 1 000 to 8 000 V for 8 h to give a total of 180 000 Vh. All IEF steps were carried out at 20 °C. After the first-dimensional IEF, IPG gel strips were placed in an equilibration solution (6 mol/L urea, 200 g/L SDS, 300 g/L glycerol, 50 mol/L Tris-HCl, pH 8.8) containing 10 g/L dithiothreilol and shaken for 15 min. The gels were then transferred to the equilibration solution containing 25 g/L iodoacetamide to alkylate thiols and shaken for a further 15 min before being placed on a 125 g/L polyacrylamide gel slab. Separation in the second dimension was carried out using Tris-glycine buffer containing 1 g/L SDS, at a current setting of 5 mA/gel for the initial 0.5 h and 18 mA/gel thereafter and a temperature of 20 °C.
For silver staining, gels were immersed in ethanol: acetic acid: water (35: 7: 58) for 1.5 h, followed by washed twice in deionized water for 20 min. Gels were pretreated for 1 min in a solution of 0.2 g/L Na2S2O3 and followed by3 of 1-min washes in deionized water. Proteins were stained in a solution containing 2 g/L AgNO3 and 0.075% formalin (37 g/L formaldehyde in water) for 20 min, and washed twice in deionized water for 1 min. Subsequently, gels were developed in a solution of 0.6 g/L formaldehyde, 20 g/L Na2S2O3 and 0.004 g/L Na2S2O3. When the desired intensity was attained, the developer was discarded and reaction stopped by 10 g/L EDTA-Na2. For Coomassie Brilliant Blue staining of gels, gels were equilibrated in a solution containing 500 mL/L methanol, 50 mL/L acetic acid and 25 g/L Coomassie Brilliant Blue R-250. Gels were rinsed in 300 mL/L ethanol containing 70 mL/L acetic acid.
Protein patterns in the gels after silver staining were recorded as digitalized images using a high-resolution scanner. Gel image matching was done with PDQuest software.
Protein spots on Coomassie blue stained gel was performed essentially as described. After the completion of staining, the gel slab was washed twice with water for 10 min. The spots of interest were excised with a scalpel and put into 1.5 mL micro-tubes. The particles were washed twice with water and then twice with water/acetonitrile (1:1) for 15 min. The solvent volumes were about twice that of the gel. Liquid was removed, acetonitrile was added to the gel particles and the mixture was left for 2 h. After that, liquid was removed and the particles were rehydrated in 25 mmol/L NH4HCO3 for 5 min. Acetonitrile was added to produce a 1:1 mixture of 25 mmol/L NH4HCO3 / acetonitrile and the mixture was incubated for 15 min. All liquid was removed. Gel particles were dried in a vacuum centrifuge, reswelled in 10 mmol/L of dithiothreilol and 25 mmol/L of NH4HCO 3, and incubated for 30 min at 56 °C to reduce the peptides. After chillness of tubes to room temperature and removal of the liquid, 55 mmol/L iodoacetamide in 25 mmol/L NH4HCO3 was added. The tubes were incubated for 30 min at room temperature in the dark to S-alkylate the peptides. Then iodoacetamide solution was removed, the particles were washed with 25 mmol/L NH4HCO3 and acetonitrile, dried in a vacuum centrifuge, rehydrated in digestion buffer containing 50 mmol/L NH4HCO3 and 12.5 ng/µL trypsin (TPCK-treated, proteomics grade, Sigma, USA), incubated for 8 h - 12 h at 37 °C. After digestion, 25 mmol/L NH4HCO 3 was added, and the tube was incubated for 15 min. Acetonitrile was added and the tube was incubated for another 15 min. The supernatant was recovered, and the extraction was repeated twice with 50 g/L TFA/acetonitrile (1:1). The three extracts were pooled and dried in a vacuum centrifuge.
One µL sample with 1 µL matrix solution CCA (α -cyano-4-hydroxycinnamic acid) was spotted on the target and dried. Dried spots were analyzed in an REFLEX-III (Bluker) MALDI-TOF mass spectrometer. The spectrometer was run in positive ion mode and in reflector mode with the setting: accelerating voltage, 20 kV; grid voltage, 76%; guide wire voltage, 0.01%; and a delay time of 150 ns. The low mass gate was set at 500 m/z.
Proteins were identified by peptide mass fingerprinting with the search programs Mascot (http://http://www.matrixscience.com/ cgi/index.plpage=.1). The following search parameters were applied: SWISS-PROT and NCBI were used as the protein sequence databases, a mass tolerance of 50 ppm and one incomplete cleavage were allowed; acetylation of the N-terminus, alkylation of lysine by carboxyamidomethylation were considered as possible modifications[9].
Mini 2DE gels (7 cm, pH3-10) were used to evaluate the reproducibility of the soluble protein preparations and to quantify the protein extracts for 2DE gel analysis. To ensure quality and reproducibility of results, 2DE maps (24 cm) were established from each sample based on silver staining of at least three independent gels, pairs of gels were run simultaneously with the same power supply and subjected to subsequent image analysis. Protein extracts prepared from tissues were compared in this way and found to be highly reproducible and similar amounts of total soluble protein were yielded when analyzed on the gels.
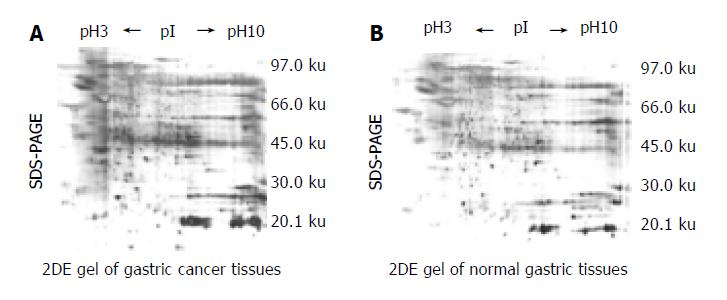
2DE Gel separation of proteins from 6 pairs of normal and cancerous epithelial cells was procured from the same gastric carcinoma specimen respectively. A series of 2DE maps were constructed for the soluble fraction proteins of human gastric tissue. A representative 2DE gel images following silver staining of cancer tissues (Figure 1A) and normal tissues ( Figure 1B) were produced in a 2DE imageMaster. Comparison of the differential protein expression between cancer and normal tissues shown in 2DE images was carried out using the AutoDetect Spots menu of PDQuest software. Figure 1A (from cancer tissues) contains a total of 356 spots, whereas Figure 1B (from normal tissues) contains a total of 382 spots. A total of 323 spots from cancer tissues could be matched to those from normal tissues. In total, we were able to identify 9 cancer-specific spots in 2DE gels. The positions of the identified proteins are shown in Figure 1A. These differences were observed in other cancerous samples. Because all of the identified spots were detectable with Coomassie Blue staining, they could be considered as abundant proteins.
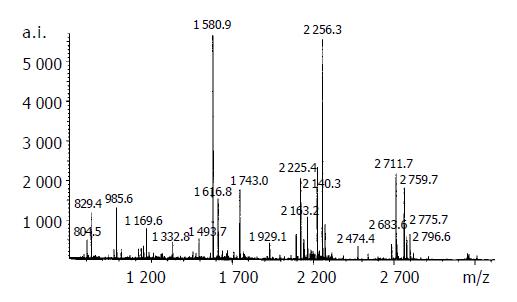
The resulting spot identification was mapped onto the analytical gels stained by Coomassie Brilliant Blue. On the map, 9 cancer-specific spots were excised and subjected to in- gel digestion followed by peptide mass fingerprinting for protein identification. Figure 2 shows the identification of the spot No. 18 as an example . We identified proteins by peptide mass fingerprinting, MS-Fit of UCSF.
The criteria used to accept identifications included the extent of sequence coverage, the number of peptides matched, the probability score, and whether human protein appeared as the top candidates in the first pass search where no restriction was applied to the species of origin.
The results of identification are summarized in the Appendix Table 1. For identified protein, probability based score greater than 73 was significant (P < 0.05). For example, in the case of annexin V, the protein score is 163. Some of these proteins, such as carbonic anhydrase I; chain B, crystal structure of fibrinogen fragment D; fibrin beta and prohibitin, have already been detected in gastric cancer tissues.
| SpotID | SWISS-PROTID | Peptidematched | Top score | TheoreticpI | TheoreticMr | Sequence coveredrate (%) | Protein name |
| 17 | gi999937 | 17/31 | 163 | 4.98 | 35 839 | 63 | Annexin V |
| 10 | gi515109 | 11/24 | 116 | 6.63 | 28 778 | 60 | Carbonic anhydrase I |
| 5 | gi2781208 | 15/43 | 105 | 5.84 | 38 081 | 55 | Chain B, crystal structure of fibrinogen fragment D |
| 12 | gi223002 | 12/33 | 92 | 7.95 | 51 358 | 42 | Fibrin beta |
| 19 | gi4505773 | 11/46 | 75 | 5.57 | 29 843 | 55 | Prohibitin |
| 4 | gi2781208 | 9/31 | 67 | 5.84 | 38 081 | 37 | Chain B, crystal structure of fibrinogen fragment D |
| 18 | gi15192925 | 8/29 | 63 | 6.90 | 31 032 | 46 | Alcohol dehydrogenase |
| 16 | gi15895617 | 9/31 | 58 | 9.01 | 42 229 | 34 | Sugar transaminase, involved in dTDP-4, 6-dideoxyglucose biosynthesis |
| 11 | gi1586816 | 10/35 | 55 | 9.33 | 42 071 | 27 | Jerky gene |
Proteome based profiling employs the measurement of protein expression pattern for the identification of individual proteins and clusters of proteins that mediate particular aspects in a physiological and pathophysiological process. The measurement of protein expression patterns of normal and disease tissues or cell populations will lead to the characterization of diagnostic and prognostic markers, and it can be further employed for the analysis of the disease stage which might also have an impact on the therapy. Thus, preferably small clusters of proteins represent the ideal diagnostic markers enabling an easier and more accurate diagnosis of diseases for better therapy[10].
This study was based on an ongoing proteomic analysis of gastric cancer aiming at screening the protein markers in the proteome for diagnosis of gastric cancer. With the availability of high-throughput 2DE gels and initial screening by using automated procedures, identification of changes in the proteome in various tissues will be possible. The approach we described in this study has shown that high-throughput analysis will be a valuable tool. An effort is currently being made using proteome based techniques[11]. 2DE pattern of normal and diseased tissues revealed a number of polypeptides associated with gastric cancer, which were expressed in gastric cancer tissues, but absent in normal gastric tissues. Some of them might possibly be identified and serve as diagnostic and prognostic markers in gastric cancer.
In the preparation of 2DE maps presented in this study, tissue samples from different individuals were used without pooling of samples. The total homogenate was fractionated by ultracentrifugation into soluble fraction[12]. In order to minimize the influence of the methodology, we attempted, when possible, to make protocols for 2DE PAGE alike. Our initial analyses of six normal tissue samples indicated the overall protein pattern remained very similar across the samples.
We identified nine protein spots which were expressed in cancerous tissues but absent in normal gastric tissues. Of the 9 position identifications, spots 4, and 5 were found as multiple spots on the 2DE gels. Those included the subunits of the proteins, both of which focused in several pI positions but had the same molecular mass. It indicated that these gene products were present as isoforms with post-translation modification[13].
Annexins are Ca2+ and phospholipid binding proteins forming an evolutionarily conserved multigene family. For some annexins, it appears that they participate in the regulation of membrane organization and membrane traffic and the regulation of ion (Ca2+) currents across membranes or Ca2+ concentrations within cells. Some members of the family have been identified extracellularly where they can act as receptors for serum proteases on the endothelium as well as inhibitors of neutrophil migration and blood coagulation[14]. Annexin V is used widely as a marker for apoptotic cells, the annexin V mutants showed defective homotypic cell adhesion and resistance to Ca 2+-dependent apoptotic agents without exhibiting any changes in the generation of cytosolic Ca2+ fluxes[15] . Annexin V also known as calphobindindin I, has been shown to be an endogenous inhibitor of protein kinase C, a key enzyme in cellular signal transduction. The inhibition of protein kinase C by annexin V is presumed to be ultimately related to carcinogenesis and studies have demonstrated that a decrease in the production of annexin V may lead to the dysregulated protein kinase C[16,17]. Annexin V has been found in other cancer tissues or cell lines[18,19], but its presence in gastric cancer has not be documented.
Carbonic anhydrase (CA) is the zinc-containing metalloenzyme that catalyzes the reversible hydration of CO2. The role of the enzyme has been thoroughly investigated. The main functions of the enzyme are to produce HCO3- for the intermediate metabolism and to maintain pH, water, and ion equilibrium in the body[20]. CAs show various levels of catalytic activity and binding to inhibitors. They have considerable diversity in tissue distribution and cellular localization, and they perform a variety of biological functions. CAI protein is associated with cell growth. It is likely expressed by rapidly proliferating tumor cells or cells that are about to enter the proliferative state, because the CA domain and other elements of the molecule take part in the regulation of cell growth in certain tumor cell types[21].
The presence of fibrin (ogen) within the tumor stroma likely affects the progression of tumor cell growth and metastasis[22]. The deposition of fibrin (ogen), along with other adhesive glycoproteins, into the extracellular matrix serves as a scaffold to support binding of growth factors and to promote the cellular responses of adhesion, proliferation, and migration during angiogenesis and tumor cell growth. Inappropriate synthesis and deposition of extracellular matrix constituents are linked to altered regulation of cell proliferation, leading to tumor cell growth and malignant transformation[23,24]. Fibrin deposition occurs within the stroma of a majority of tumor types. Fibrin (ogen) content was significantly higher in malignant tumor patients than that in benign disease patients, significant reduction was observed after treatment and became elevated again when there was recurrence or metastasis[25]. Biggerstaff et al[26] suggested that coagulation activation and the subsequent increase in circulating fibrin may enhance platelet adhesion to circulating tumor cells and thereby facilitate metastatic spread. Assessment of fibrin (ogen) not only helps to diagnose cancers but also evaluates the therapautic effect and prognosis[27].
Prohibitin proteins have been implicated in cell proliferation, ageing and the maintenance of mitochondria integrity[28], prohibitins are present in the inner mitochondrial membrane and always bound to each other. They are expressed during development and their expression levels are indicative of a role in mitochondrial metabolism[29]. High level expression of the proteins is consistently seen in primary human tumors. The prohibitin protein has been found having various functions, including cell cycle regulation, apoptosis, assembly of mitochondrial respiratory chain enzymes and ageing. We are currently trying to identify the additional proteins whose expression is significantly altered in cancerous tissue.
This study also confirms that proteomic analysis is a powerful tool for the discovery of such molecular markers. Proteomic analysis allows the characterization of picoquantities of proteins with MS and changes in the levels inherent to the pathophysiology of any cell type, tissue, or whole organism. It is hoped that the identification of protein markers by this approach could discriminate cancerous from normal cells. As demonstrated here, proteomic analysis may be efficiently used to identify new indicators for the diagnosis and prognosis of cancer progression.
In conclusion, the differences of the proteins between normal gastric epithelial cell and malignant cells are complex. Our data only show a few of the highly expressed spots. Further basic and clinical investigation will be needed to demonstrate if these proteins could be markers for GC and to evaluate other non-constant spots in relation to the clinical condition of the patient.
The authors would like to thank Dr. Bo-Qing Li and Qing-Hua Zhou at Chinese Center for Disease Control and Prevention for their assistance in the experiments.
| 1. | Lawrie LC, Fothergill JE, Murray GI. Spot the differences: proteomics in cancer research. Lancet Oncol. 2001;2:270-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 53] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 2. | Zhang M, Martin KJ, Sheng S, Sager R. Expression genetics: a different approach to cancer diagnosis and prognosis. Trends Biotechnol. 1998;16:66-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 3. | Torrado J, Plummer M, Vivas J, Garay J, Lopez G, Peraza S, Carillo E, Oliver W, Muñoz N. Lewis antigen alterations in a population at high risk of stomach cancer. Cancer Epidemiol Biomarkers Prev. 2000;9:671-674. [PubMed] |
| 4. | Xu CT, Pan BR, Zhang LZ, Li XX, Wang J. Significance of serum tumor markers CA50 and CEA in gastric cancer. China J New Gastroenterol. 1996;2:16-19. |
| 5. | Macgregor PF, Squire JA. Application of microarrays to the analysis of gene expression in cancer. Clin Chem. 2002;48:1170-1177. [PubMed] |
| 6. | Gygi SP, Corthals GL, Zhang Y, Rochon Y, Aebersold R. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc Natl Acad Sci USA. 2000;97:9390-9395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 946] [Cited by in RCA: 850] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 7. | Ni XG, Zhao P, Liu Y, Zhao XH. [Application of proteomic approach for solid tumor marker discovery]. Ai Zheng. 2003;22:664-667. [PubMed] |
| 8. | Emmert-Buck MR, Gillespie JW, Paweletz CP, Ornstein DK, Basrur V, Appella E, Wang QH, Huang J, Hu N, Taylor P. An approach to proteomic analysis of human tumors. Mol Carcinog. 2000;27:158-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 9. | Görg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, Weiss W. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 2000;21:1037-1053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 10. | Yu YL, Yang PY, Fan HZ, Huang ZY, Rui YC, Yang PY. Protein expressions in macrophage-derived foam cells: comparative analysis by two-dimensional gel electrophoresis. Acta Pharmacol Sin. 2003;24:873-877. [PubMed] |
| 11. | Bécamel C, Galéotti N, Poncet J, Jouin P, Dumuis A, Bockaert J, Marin P. A proteomic approach based on peptide affinity chromatography, 2-dimensional electrophoresis and mass spectrometry to identify multiprotein complexes interacting with membrane-bound receptors. Biol Proced Online. 2002;4:94-104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 12. | Mikami S, Kishimoto T, Hori H, Mitsui T. Technical improvement to 2D-PAGE of rice organelle membrane proteins. Biosci Biotechnol Biochem. 2002;66:1170-1173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 13. | Steel LF, Shumpert D, Trotter M, Seeholzer SH, Evans AA, London WT, Dwek R, Block TM. A strategy for the comparative analysis of serum proteomes for the discovery of biomarkers for hepatocellular carcinoma. Proteomics. 2003;3:601-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 119] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 14. | Katayanagi K, Van de Water J, Kenny T, Nakanuma Y, Ansari AA, Coppel R, Gershwin ME. Generation of monoclonal antibodies to murine bile duct epithelial cells: identification of annexin V as a new marker of small intrahepatic bile ducts. Hepatology. 1999;29:1019-1025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 15. | Nimmo MC, Carter CJ. The antiphospholipid antibody syndrome: A riddle wrapped in a mystery inside an enigma. Clinical Applied Immunol Rev. 2003;4:125-140. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 16. | Karube A, Shidara Y, Hayasaka K, Maki M, Tanaka T. Suppression of calphobindin I (CPB I) production in carcinoma of uterine cervix and endometrium. Gynecol Oncol. 1995;58:295-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Shibata S, Sato H, Ota H, Karube A, Takahashi O, Tanaka T. Involvement of annexin V in antiproliferative effects of gonadotropin-releasing hormone agonists on human endometrial cancer cell line. Gynecol Oncol. 1997;66:217-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 18. | Seow TK, Ong SE, Liang RC, Ren EC, Chan L, Ou K, Chung MC. Two-dimensional electrophoresis map of the human hepatocellular carcinoma cell line, HCC-M, and identification of the separated proteins by mass spectrometry. Electrophoresis. 2000;21:1787-1813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 19. | Xin W, Rhodes DR, Ingold C, Chinnaiyan AM, Rubin MA. Dysregulation of the annexin family protein family is associated with prostate cancer progression. Am J Pathol. 2003;162:255-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 144] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 20. | Ivanov S, Liao SY, Ivanova A, Danilkovitch-Miagkova A, Tarasova N, Weirich G, Merrill MJ, Proescholdt MA, Oldfield EH, Lee J. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol. 2001;158:905-919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 509] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 21. | Nógrádi A. The role of carbonic anhydrases in tumors. Am J Pathol. 1998;153:1-4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 22. | Palumbo JS, Kombrinck KW, Drew AF, Grimes TS, Kiser JH, Degen JL, Bugge TH. Fibrinogen is an important determinant of the metastatic potential of circulating tumor cells. Blood. 2000;96:3302-3309. [PubMed] |
| 23. | Stewart DA, Cooper CR, Sikes RA. Changes in extracellular matrix (ECM) and ECM-associated proteins in the metastatic progression of prostate cancer. Reprod Biol Endocrinol. 2004;2:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 99] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 24. | Sahni A, Odrljin T, Francis CW. Binding of basic fibroblast growth factor to fibrinogen and fibrin. J Biol Chem. 1998;273:7554-7559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 173] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 25. | Holmbeck K, Bianco P, Birkedal-Hansen H. MT1-mmp: a collagenase essential for tumor cell invasive growth. Cancer Cell. 2003;4:83-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 26. | Biggerstaff JP, Seth NB, Meyer TV, Amirkhosravi A, Francis JL. Fibrin monomer increases platelet adherence to tumor cells in a flowing system: a possible role in metastasis. Thromb Res. 1998;92:S53-S58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 27. | Kim HK, Lee KR, Yang JH, Yoo SJ, Lee SW, Jang HJ, Park SJ, Moon YS, Park JW, Kim CM. Plasma levels of D-dimer and soluble fibrin polymer in patients with hepatocellular carcinoma: a possible predictor of tumor thrombosis. Thromb Res. 2003;109:125-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 28. | McClung JK, Jupe ER, Liu XT, Dell'Orco RT. Prohibitin: potential role in senescence, development, and tumor suppression. Exp Gerontol. 1995;30:99-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 145] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 29. | Nijtmans LG, de Jong L, Artal Sanz M, Coates PJ, Berden JA, Back JW, Muijsers AO, van der Spek H, Grivell LA. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 2000;19:2444-2451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 447] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
Edited by Zhu LH Proofread by Chen WW and Xu FM