Published online Jul 15, 2004. doi: 10.3748/wjg.v10.i14.2103
Revised: February 10, 2004
Accepted: March 2, 2004
Published online: July 15, 2004
AIM: To obtain the entire gene open reading frame (ORF) and to construct the expression vectors for recombinant allergen production.
METHODS: Gene fragments corresponding to the gene specific region and the cDNA ends of pollen allergens of short ragweed (Rg, Ambrosia artemisiifolia L.) were obtained by pan-degenerate primer-based PCR and rapid amplification of the cDNA ends (RACE), and the products were mixed to serve as the bridging PCR (BPCR) template. The full-length gene was then obtained. Partially overlapping primer-based PCR (POP-PCR) method was developed to overcome the other problem, i.e., the non-specific amplification of the ORF with routine long primers for expression insert decoration. Northern blot was conducted to confirm pollen sources of the gene. The full-length coding region was evaluated for its gene function by homologue search in GenBank database and Western blotting of the recombinant protein Amb a 8 (D106) expressed in Escherichia coli pET-44 system.
RESULTS: The full-length cDNA sequence of Amb a 8(D106) was obtained by using the above procedure and deduced to encode a 131 amino acid polypeptide. Multiple sequence alignment exhibited the gene D106 sharing a homology as high as 54%-89% and 79%-89% to profilin from pollen and food sources, respectively. The expression vector of the allergen gene D106 was successfully constructed by employing the combined method of BPCR and POP-PCR. Recombinant allergen rAmb a 8(D106) was then successfully generated. The allergenicity was hallmarked by immunoblotting with the allergic serum samples and its RNA source was confirmed by Northern blot.
CONCLUSION: The combined procedure of POP-PCR and BPCR is a powerful method for full-length allergen gene retrieval and expression insert decoration, which would be useful for recombinant allergen production and subsequent diagnosis and immunotherapy of pollen and food allergy.
- Citation: Tao AL, He SH. Bridging PCR and partially overlapping primers for novel allergen gene cloning and expression insert decoration. World J Gastroenterol 2004; 10(14): 2103-2108
- URL: https://www.wjgnet.com/1007-9327/full/v10/i14/2103.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i14.2103
Atopic diseases such as asthma, rhinitis, eczema, pollen allergies and food allergies have been increasing in most industrialized countries of the world during the last 20 years[1]. In the past years, we have improved our ability to recognize certain aspects of the pathogenesis of inflammatory bowel diseases, but our ability to effectively treat food and pollen allergy remains limited. The limitations for developing effective treatment regimens are due to some still unresolved and ambiguous aspects of the cross-reactivity of food and pollen allergy[2]. Among the “cross-reactivity” food allergies, the so-called pollen-related food allergies are the most important. Patients with pollinosis have a high risk of developing a related food allergy, up to 70% of the patients are also allergic to fruits, vegetables or nuts[3,4]. Short ragweed (Rg, Ambrosia artemisiifolia L.) is an important source of airborne allergens all over the world and has become an immunodominant allergen in China. Allergen-specific immunotherapy (SIT) represents one of the few curative approaches towards allergy[5], for which a powerful method is to express the recombinant allergen proteins in vitro. Therefore, it is pivotal to obtain full-length allergen gene and to construct the expression vector. Generally, many full-length genes are acquired by prior acquisition of the specific fragments, and subsequently constructed in conception with computers by joining the amphi-end sequences obtained by rapid amplification of cDNA end (RACE)[6]. In the process, a frequently occurred problem is the existence of alternatives in the amplification template. That is to say, though the length of amplified sequences is consistent with anticipation, the nucleotide compositions in different sequences are dramatically different and the existence of polymorphism is usually observed[7-11]. Screening and detection of different clones obtained would account for the main expenditure of the prophase work of gene cloning. As for the expression vector construction, specific primers needed usually comprise two parts. One is the region for restriction recognition or the vector specific sequences for ligation-independent cloning (LIC), like newly developed pET-44 EK/LIC system. The other is the gene specific region for stringent polymerase chain reaction (PCR) . The melting temperature (Tm) of such primers always goes beyond the ordinary limit of 72 °C, and the amplification specificity hence is reduced. It would be more disturbing when the GC content of the open reading frame (ORF) ends greatly deviates from 50%, which would usually occur in the 5’ end of the ORF. To obviate the aforementioned problems, we have developed the partially overlapping primer-PCR (POP-PCR) method to surmount the non-specificity problem for long primers. This was then combined with bridging PCR (BPCR), which was coined for the PCR assay on joining two fragments containing a region of sequence similarity[12,13] , to conquer the difficulty in retrieving the full-length gene in cDNA pool that was originally used for gene fragments cloning. In the current study, we successfully constructed the expression vector and expressed the allergen protein from Rg pollens, and hallmarked the allergenicity on the purified fusion protein.
Eight selected sera of patients with a well-defined case history of Rg pollen allergy and specific IgE antibodies against Rg pollen allergen (higher than 3.5 kUA/L) were taken in Allergy Clinics, Shenyang North Hospital, Shenyang, China and used in this study. Thirteen sera from non-allergic individuals were recruited from the First Affiliated Hospital of Medical College, Shantou University, Shantou, China and used as negative controls.
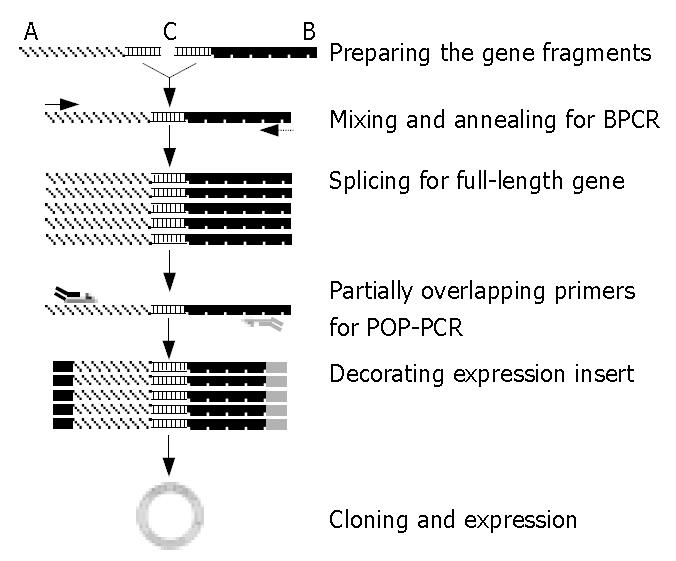
Pollens of Rg were harvested from Wuhan, one of the four pervasive areas of short ragweed in China, and immediately submerged in liquid nitrogen. Following powder-grinding, total RNA was extracted by using RNeasy Maxi kit (Qiagen Valencia, CA, USA) according to the manufacturer’s instructions. Reverse transcription (2 μg total RNA per sample) was then performed by using ProtoScript first strand cDNA synthesis kit (New England Biolabs, Beverly, MA, USA) according to the manufacturer’s instructions. Subsequently, the 5’ end of the gene (gene fragment A, Figure 1) was obtained by RT-PCR with primer pair Sg1P5/S3D10 (Table 1). Whereas the 3’ end (gene fragment B in Figure 1) was acquired by RACE (rapid amplification of the cDNA end) method[6]. The cDNA pool, along with fragment A and fragment B would be used for the subsequent PCR templates.
| Code | Definition | Sequence (5’→3’) | Tm (°C) |
| Sg1P5 | For fragment A and BPCR | ATGTCGTGGCAGGCGTACGT | 66.0 |
| S3D10 | For fragment A | GGACAATGCAACATGCTTGTTGAGAGG | 70.4 |
| S5D106 | For fragment B | GTGGAAAGAAGGGAGCAGGAGG | 66.4 |
| RAC3 | For fragment B and BPCR | GCTGTCAACGATACGCTACGTAACGG | 68.4 |
| DEN3 | For ORF of D106 | TAATTAGAAACCCTGTTCGAGGAGATAGTCACC | 68.4 |
| POP51 | For POP-PCR | GACGACGACAAGATGTCGTGGCAGGCGTACGT | 80.8 |
| POP52 | For POP-PCR | GACGACGACAAGATGTCGTGGCA | 69.9 |
| POP31 | For POP-PCR | GAGGAGAAGCCCGGTTTACATGCCCTGATCGATGAGAT | 80.7 |
| POP32 | For POP-PCR | GAGGAGAAGCCCGGTTTACATGCC | 69.7 |
The amplification products A and B were mixed in equal volume and diluted 15-fold in distilled water. Bridging PCR (BPCR) outlined in Figure 1 was carried out by using 5 μL of the above diluents as template in 25 μL of the reaction mixture containing 1.25 U HotstarTaq DNA polymerase (Qiagen), 1 × PCR buffer (Qiagen), 0.2 mmol/L dNTP, 0.4 μmol/L each of the primer pair Sg1P5 and RAC3 (Table 1). Thermal cycling was performed in a Peltier thermal cycler PTC-200 (MJ Research, Watertown, MA, USA) as follows. After an initial step at 95 °C for 15 min as recommended to activate the enzyme, the reaction temperature was decreased to 66 °C at the rate of -0.1 °C/s followed by an extension at 72 °C for 5 min, then 7 touchdown cycles were carried out at 94 °C for 45 s, at 68 °C for 30 s and at 72 °C for 30 s, with the annealing temperature decreased by 1 °C per cycle. Subsequently, the reaction mixture was subjected to 30 conventional cycles at 94 °C for 45 s, at 66 °C for 30 s, and at 72 °C for 2 min. Conventional PCR was also conducted as a control by using cDNA pool aliquot as template. As visualized on a 12 g/L agarose gel, the target bands were recovered with a QIAquickTM gel extraction kit (Qiagen), cloned into pGEM-T easy vector (Promega Company, Madison, WI, USA), and transformed into E. coli strain JM109 (Promega). Sequencing of the positive clones was performed by BioAsia Biotechnology Co., Ltd (Shanghai, China). Sequence BLAST was performed in GenBank database.
To decorate the expression insert, primers were designed according to the sequence of the gene-coding region and the specific sequence of the expression vector as recommended by the manufacturer’s instructions. The melting temperature (Tm) of all primers was calculated by using the software Omiga 2.0 (Oxford Molecular Ltd, Oxford, UK). Shorter overlapping primers were additionally designed for the long primers with the Tm beyond 72 °C (Table 1). The sequences of this kind of primers each were identical to the 5’ terminal sequence of their corresponding longer ones. Pooling together the shorter and longer ones in different molar ratio formed the partially overlapping primers (Figure 1). Pilot assays were conducted to optimize the molar ratio of the two primers in the partially overlapping primer pairs for PCR. The optimal proportion was subsequently chosen for partially overlapping primer PCR (POP-PCR), by using the product recovered from BPCR as a template. As a control, the cDNA pool, originally used for fragment gene cloning, was also used as the PCR template in parallel. The PCR mixture and the initial step were similar to those for BPCR, but with some changes in denaturing temperature and extension time. Following the initial denaturing step, 12 touchdown cycles at the initiative annealing step at 72 °C for 45 s, and an extension at 72 °C for 5 min were performed, followed by 30 extenuation cycles, which were initiated at 95 °C for 45 s, at 72 °C for 1 min and at 72 °C for 30 s, with the annealing time decreased by 1 s per cycle, and ended by a final extension at 72 °C for 10 min. The annealing temperature was usually adjusted according to the Tm of the primer being used in the reaction. The target bands visualized on the gel were purified and sequenced as described above. However, the plasmids were cloned into E. coli strain NovaBlue SinglesTM cells (Novagen, Madison, WI, USA), according to the manufacturer’s instruction.
Northern blot was performed according to the manufacture’s manual with a slight modification. Briefly, 20 μg RNAs from pollen and the mixture of calyx and pedicel was loaded on a formadehyde-containing gel. After running at 40 volts for 3 h or so, the RNA bands were electrically transferred onto Nytran SuPerCharge nylon transfer membranes (Schleicher & Schull BioScience, Inc. New Hampshire, USA). Before stained with methylene blue [0.2 g/L in 0.3 mol/L sodium acetate (pH5.5)] to investigate the transferring efficiency, RNA was UV cross-linked onto the membrane. After destaining, BrightStarTM Psoralen-biotin (Ambion Inc., Austin, TX, USA) labeled probe D106 was applied for hybridization in the ULTRhybTM hybridization buffer (Ambion Inc., USA). Subsequent washing was performed twice with a buffer containing 2 × SSC (0.3 mol/L NaCl, 0.03 mol/L sodium citrate) and 1 g/L SDS, before its probe was detected by Phototope star detection kit (Ambion). Finally, the filter was exposed to Fuji medical X-ray film at room temperature for several min.
After confirmation of the in-frame insertion, the positive clones were incubated overnight at 37 °C on an orbital shaker. The plasmids therein were extracted by using QIAprep Miniprep kit (Qiagen) and transformed into E. coli strain BL21(DE3) (Novagen) for expression according to the manufacturer’s instructions with a minor modification. Following overnight proliferation of the expression transformants, the cells were inoculated in liquid LB medium at the rate of 1%. After a further incubation for 4 h or so, expression of protein D106 was induced with 0.5 mmol/L isopropyl-D-thio-galactopyranoside (IPTG) for 2 h at 30 °C before harvest. The expression results were analyzed on 150 g/L SDS-PAGE.
After induced expression was confirmed, the recombinant allergen Amb a 8(D106) was purified with S·Tag thrombin purification kit (Novagen). Fifteen micrograms of fusion protein was resolved on 150 g/L SDS-PAGE and electrically transferred to PVDF membrane (Amersco, Solon, Ohio, USA). After the membrane was blocked overnight with 40 g/L BSA solution, Rg allergic serum (diluted 1:7) was added to each membrane strip for 8 h before addition of peroxidase conjugated goat anti-human IgE (Sigma-Aldrich, Inc., Saint Louis, MO, USA). The blotted band was visualized with DAB chromogenic substrate solution[14] (Amersco).
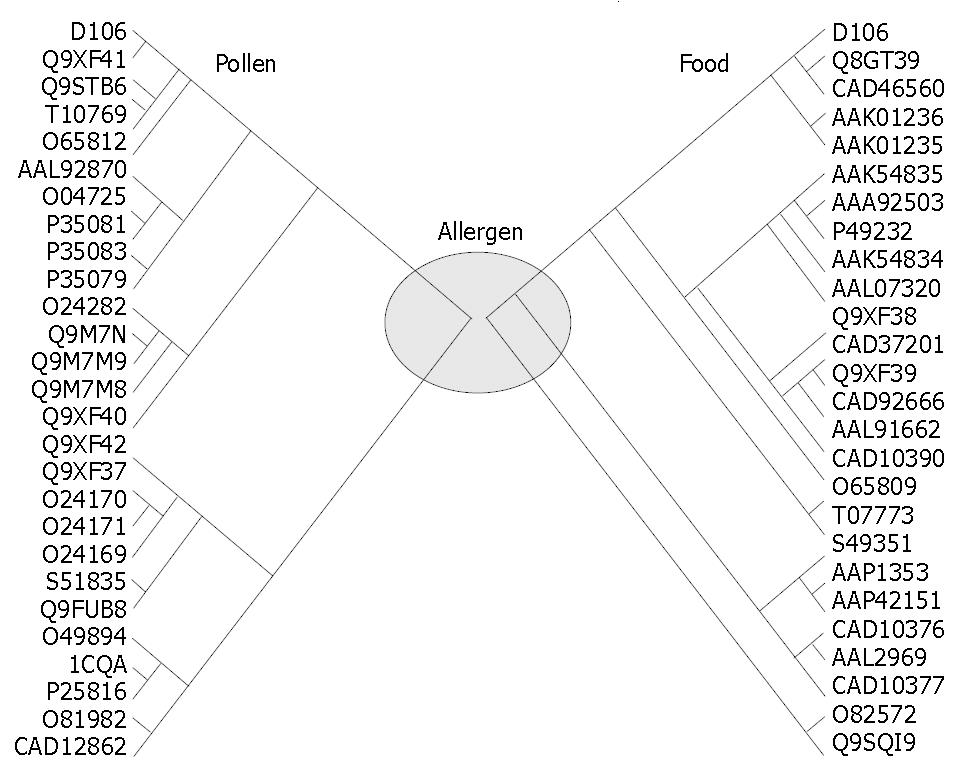
Gene fragments A and B corresponding to the 5’- and 3’-ends of the gene were enriched by RT-PCR and subsequently inserted into the vector. The sequence analysis demonstrated that fragments A and B, sized 363 bp and 431 bp respectively, contained a common region of 126 bp, which would provide the junction region for BPCR. The theoretical full-length gene joined contained the stop codon, poly (A) tail and primer sequences (Figure 2). Bridging the two fragments produced a novel theoretical full-length gene, which can be available under GenBank Accession No AY268426. BLAST with the amino acid sequence deduced from the full-length cDNA sequence of D106 retrieved a large number of homologous proteins corresponding to pollen, food and contact allergens from different species, all of which belonged to profilin family. The BLAST data also showed that Amb a 8(D106) shared a homology of 54%-89% with pollen allergens and of 79%-89% with food allergens, which could help to cluster Amb a 8(D106) into both pollen and food allergen groups with short distances (Figure 3).
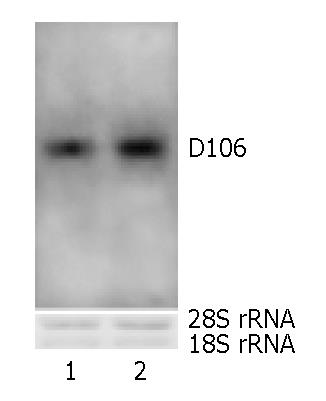
To confirm the RNA derivation of cDNA of D106, Northern blot was performed. After blotting and UV-cross linking, methylene blue-staining of the membrane exhibited the sharp rRNA bands, suggesting the successful transferring and good quality of RNA. The sharp hybridization bands proved the pollen derivation of the cloned allergen cDNA (Figure 4). D106 could be detected both from pollen RNA and the RNA mixture from calyx and pedicel, which ruled out the PCR false derivation of D106.
In order to clone the full-length gene coding region for expression vector construction, we first resorted to general PCR by using the cDNA pool as a template with the primers formerly used. However, no sharp bands of expected sizes were exhibited on the loaded agarose gel for general PCR, though a number of PCR conditions were applied. After the template was changed to the mixture of gene fragments A and B previously acquired, the bridging PCR (BPCR), as outlined in Figure 1, produced the optimal results with sharp bands and exact size observed (Figure 5). The gene sequence achieved was identical to that spliced from gene fragments A and B, indicating that the ORF of the target gene D106 was contained.
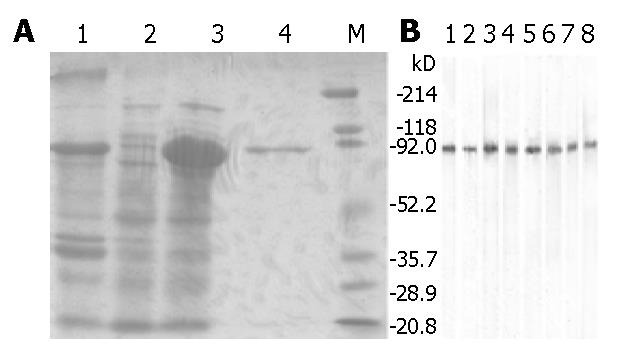
For comparison between conventional megaprimers and partially overlapping primers (POP), their amplification products were exhibited in parallel on the agarose gel (Figure 5). Longer primers and their relative shorter ones for POP-PCR listed in Table 1 were pre-mixed in a molar ratio of 1:1, 1:10, 1:50, 1:100. Hence 16 POP-pairs in different molar ratios were formed corresponding to both ends of the ORF of the gene, each of them was used for one reaction only. Comparison of the parallel PCR results showed that forward and reverse POPs in the molar ratios of 1:10 and 1:1 could produce the optimal amplification results, while conventional PCR exhibited the smear products with wrong sizes (Figure 5) . The products recovered from target bands were successfully introduced into the expression vector pET-44 EK/LIC (Novagen) and transformed into E. coli strain NovaBlue SinglesTM competent cells. Sequencing of the positive clones demonstrated that the insert was in-frame and identical to those splices from gene fragments A and B and flanked by the vector specific sequence at both sides, indicating that the insertion was correct.
Following proliferation of the clones selected, plasmids were extracted and successfully transformed into E. coli strain BL21 (DE3) for expression. SDS-PAGE analysis showed that the fusion protein was expressed mainly in a soluble form, with a molecular weight of 70 kDa as expected (Figure 6A). Immunobotting result showed that the purified fusion protein could bind to IgE in the sera of allergic patients (Figure 6B), suggesting the allergenicity and the correct expression of the allergen protein.
As industrialization developed, atopic diseases have been increasingly concerned in the past decades[1]. It is known that pollen-allergic patients frequently present allergic symptoms after ingestion of several kinds of plant-derived foods. The majority of these reactions were caused by cross-reactive structures that are present in pollens[2]. Allergic proteins containing these structures are often called ‘panallergens’. Cloning the panallergen gene and expressing the recombinant protein would be one of the fewer prospective ways to pollen-food allergy diagnosis and immunotherapy[15]. This study aiming this goal successfully cloned an allergen gene D106 from short ragweed pollens and expressed the recombinant protein. Interestingly, the newly cloned gene showed a high homology not only with other pollen-derived profilins but also with food-derived profilins from numerous species. The allergenicity of the recombinant protein was primarily detected by its binding to the serum IgE from the Rg pollen allergic patients, of which 20% of the chief complaints expressed the food allergy of different kinds. Based on the homology results and the widely accepted rule-of-thumb that 30% or 35% identity over aligned regions could suffice for the structural or functional deduction[4,16,17], it is therefore tempting to speculate that the recombinant protein acquired in this study would be a useful tool both for pollen and food allergy study, and for diagnosis and immunotherapy.
The novel gene discovery by using suppression and subtractive hybridization (SSH)[18,19] and/or reverse genetic method[20] could clearly validate the overall strategy for the preparation of gene fragments. However, obtaining the full-length genes is needed for more investigations of gene functions. One of the pivotal steps in the process of obtaining the full-length genes is to generate the target genes from their fragments. The most commonly used methods for this at present were to screen the cDNA library[21-23], or back to the cDNA pool that was originally used for gene fragment detection[23]. Numerous polymorphic clones obtained would render these methods to time-consuming, expensive and liable to produce the microherterogeneity of the genes. In BPCR, only if an appropriate length region of sequence similarity existed, the two fragments could be spliced together. Obviously, the spliced sequence would be faithfully identical to that of the fragments originally used. Therefore no more screening work was needed. In order to anneal the two fragments and form a template molecule, it is a key point to optimize the annealing temperature according to the Tm of the junction regions of two gene fragments to be spliced, with the primers used in the reaction. Following this principle, BPCR would splice any two gene fragments with homology junction. Notably for the junction sequences either coming from the nature, or intentionally synthesized (see below), exact identity is not necessary but it does require a certain degree of homology to anneal the two fragment genes. Therefore, two heterogeneity genes could be spliced by BPCR method to form a novel chimeric gene or hybrid gene, which would be useful for gene function study and disease treatment.
Introducing restriction site and/or the specific sequences of vector to the primers is considered as a crucial step for exact insertion of exogenous gene into the vector, through which relatively long primers could be produced. It was well known that these longer primers would be easier to enhance the chance of non-specific amplification. To eliminate the potential non-specific amplification, partially overlapping primer technique was developed in the current study. According to the outward overhang sequence of the longer primers, the shorter identical sequence was synthesized and mixed in an appropriate molar ratio with the longer ones, thus forming the partially overlapping primer mixture. With this technique, increased amplification specificity was achieved, and the tedious screening process of target clones was avoided. According to the protocol of the partially overlapping primer, POP-PCR can be used for the amplification assays that are relevant to the addition of a length of extending sequence. It is therefore tempting to suggest that any genes that have no sequence similarities can be accommodated with the junction sequence and spliced together with repeated use and reciprocal combination of BPCR and POP-PCR to form a novel gene (or full-length gene). As this is concerned, a relatively more complicated procedure but with different efficiencies and proposes has been recently developed[24].
| 1. | Miescher SM, Vogel M. Molecular aspects of allergy. Mol Aspects Med. 2002;23:413-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 43] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 2. | Vieths S, Scheurer S, Ballmer-Weber B. Current understanding of cross-reactivity of food allergens and pollen. Ann N Y Acad Sci. 2002;964:47-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 296] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 3. | Lorenz AR, Scheurer S, Haustein D, Vieths S. Recombinant food allergens. J Chromatogr B Biomed Sci Appl. 2001;756:255-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 4. | Brusic V, Petrovsky N, Gendel SM, Millot M, Gigonzac O, Stelman SJ. Computational tools for the study of allergens. Allergy. 2003;58:1083-1092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 5. | Spangfort MD, Mirza O, Ipsen H, Van Neerven RJ, Gajhede M, Larsen JN. Dominating IgE-binding epitope of Bet v 1, the major allergen of birch pollen, characterized by X-ray crystallography and site-directed mutagenesis. J Immunol. 2003;171:3084-3090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 133] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 6. | Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998-9002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3036] [Cited by in RCA: 3393] [Article Influence: 89.3] [Reference Citation Analysis (0)] |
| 7. | Kruse S, Kuehr J, Moseler M, Kopp MV, Kurz T, Deichmann KA, Foster PS, Mattes J. Polymorphisms in the IL 18 gene are associated with specific sensitization to common allergens and allergic rhinitis. J Allergy Clin Immunol. 2003;111:117-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 100] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 8. | Karjalainen J, Hulkkonen J, Pessi T, Huhtala H, Nieminen MM, Aromaa A, Klaukka T, Hurme M. The IL1A genotype associates with atopy in nonasthmatic adults. J Allergy Clin Immunol. 2002;110:429-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 9. | Cookson W. Genetics and genomics of asthma and allergic diseases. Immunol Rev. 2002;190:195-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 73] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Vailes LD, Sun AW, Ichikawa K, Wu Z, Sulahian TH, Chapman MD, Guyre PM. High-level expression of immunoreactive recombinant cat allergen (Fel d 1): Targeting to antigen-presenting cells. J Allergy Clin Immunol. 2002;110:757-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Hales BJ, Hazell LA, Smith W, Thomas WR. Genetic variation of Der p 2 allergens: effects on T cell responses and immunoglobulin E binding. Clin Exp Allergy. 2002;32:1461-1467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Mehta RK, Singh J. Bridge-overlap-extension PCR method for constructing chimeric genes. Biotechniques. 1999;26:1082-1086. [PubMed] |
| 13. | Liu S, Thaler DS, Libchaber A. Signal and noise in bridging PCR. BMC Biotechnol. 2002;2:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 14. | Reindl J, Rihs HP, Scheurer S, Wangorsch A, Haustein D, Vieths S. IgE reactivity to profilin in pollen-sensitized subjects with adverse reactions to banana and pineapple. Int Arch Allergy Immunol. 2002;128:105-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 41] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Ferreira F, Wallner M, Breiteneder H, Hartl A, Thalhamer J, Ebner C. Genetic engineering of allergens: future therapeutic products. Int Arch Allergy Immunol. 2002;128:171-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 39] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 16. | Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1208] [Cited by in RCA: 1192] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 17. | Hileman RE, Silvanovich A, Goodman RE, Rice EA, Holleschak G, Astwood JD, Hefle SL. Bioinformatic methods for allergenicity assessment using a comprehensive allergen database. Int Arch Allergy Immunol. 2002;128:280-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 120] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 18. | Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025-6030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2262] [Cited by in RCA: 2004] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 19. | Sers C, Tchernitsa OI, Zuber J, Diatchenko L, Zhumabayeva B, Desai S, Htun S, Hyder K, Wiechen K, Agoulnik A. Gene expression profiling in RAS oncogene-transformed cell lines and in solid tumors using subtractive suppression hybridization and cDNA arrays. Adv Enzyme Regul. 2002;42:63-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Yu CJ, Lin YF, Chiang BL, Chow LP. Proteomics and immunological analysis of a novel shrimp allergen, Pen m 2. J Immunol. 2003;170:445-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 188] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 21. | Ding H, Griesel C, Nimtz M, Conradt HS, Weich HA, Jäger V. Molecular cloning, expression, purification, and characterization of soluble full-length, human interleukin-3 with a baculovirus-insect cell expression system. Protein Expr Purif. 2003;31:34-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 22. | Erez N, Milyavsky M, Goldfinger N, Peles E, Gudkov AV, Rotter V. Falkor, a novel cell growth regulator isolated by a functional genetic screen. Oncogene. 2002;21:6713-6721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Forestier M, Bänninger R, Reichen J, Solioz M. Betaine homocysteine methyltransferase: gene cloning and expression analysis in rat liver cirrhosis. Biochim Biophys Acta. 2003;1638:29-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 24. | Gao X, Yo P, Keith A, Ragan TJ, Harris TK. Thermodynamically balanced inside-out (TBIO) PCR-based gene synthesis: a novel method of primer design for high-fidelity assembly of longer gene sequences. Nucleic Acids Res. 2003;31:e143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 91] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
Edited by Wang XL Proofread by Xu FM