Published online Jun 1, 2004. doi: 10.3748/wjg.v10.i11.1625
Revised: August 4, 2002
Accepted: August 31, 2002
Published online: June 1, 2004
AIM: To compare the gene expression between La (NO3) 3-exposed and control rats in vivo.
METHODS: Rats were fed La (NO3) 3 once daily at a dose of 20 mg/kg for one month by gavage. Gene expression of hepatocytes was detected using mRNA differential display (DD) technique and cDNA microarray and compared between treated and control groups.
RESULTS: Six differentially expressed sequence tags were cloned by DD, of which five were up regulated and one was down regulated in treated rats. Two sequences were determined. One band was novel. The other shared 100% sequence homology with AU080263 Sugano mouse brain mncb Mus musculus cDNA clone MNCb-5435 5’. With DNA microarray, 136 differentially expressed genes were identified including 131 over-expressed genes and 5 under-expressed genes. Most of these differentially expressed genes were cell signal and transmission genes, genes associated with metabolism, protein translation and synthesis.
CONCLUSION: La (NO3) 3 could change the expression levels of some kinds of genes. Further analysis of the differentially expressed genes would be helpful for understanding the wide biological effect spectrum of rare earth elements.
- Citation: Zhao H, Hao WD, Xu HE, Shang LQ, Lu YY. Gene expression profiles of hepatocytes treated with La (NO3) 3 of rare earth in rats. World J Gastroenterol 2004; 10(11): 1625-1629
- URL: https://www.wjgnet.com/1007-9327/full/v10/i11/1625.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i11.1625
Rare earth (RE) includes 17 elements. According to the physical and chemical nature, 16 elements except for scandium (Sc) are categorized into two groups. One is called light RE, which is represented by cerium, including lanthanum (La), cerium (Ce), praseodymium (Pr), neodymium (Nd), promethium (Pm), samarium (Sm) and europium (Eu). The other is called heavy RE, which is represented by yttrium, including gadolinium (Gd), terbium (Tb), dysprosium (Dy), holmium (Ho), erbium (Er), thulium (Tm), ytterbium (Yb), lutetium (Lu) and yttrium (Y). Recently, RE has become one of the common xenobiotics in our surroundings as it is widely used in industry, stockbreeding and medicine, especially as trace fertilizers in agriculture, and they can be concentrated by food chain. Therefore, understanding the effects of RE on health has become more and more important.
Studies on RE toxicology have lasted for a long time. However, deeper exploration of its mechanism is highly needed. It is well known the biological effect spectrum of RE is wide and the dose-response relationship is complicated. For example, RE elements could induce chromosome damage of blood lymphocytes[1] and liver damage[2-4], depress learning and memory[5], increase or suppress cell-mediated immunity of the spleen[6], inhibit gap junctional intercellular communication[7], etc. However, safety evaluation of RE is not easy, and the research of biomarkers by molecular biology is in its infancy. It is necessary to find more sensitive biomarkers by new techniques and methods, and to acquire a deeper understanding of the mechanism.
In this paper, the molecular mechanism of RE toxicity was explored by DD and cDNA microarray. La (NO3) 3, which is the major component in RE trace fertilizers, was selected as the study material.
In the study of DD, 86-week-old Wistar rats were chosen. One half of them were male, and the others were female. To guarantee the treated rats and control had similar heredity backgrounds, the rule of same-breeding and same-sex control was applied. A La (NO3) 3-treated rat and its control were called a pair of test animals. There were two pairs of male rats and female rats, respectively. Treated rats were fed La (NO3) 3 once daily at a dose of 20 mg/kg for one month by gavage. Control rats were fed distilled water. One pair of female rats was chosen in the study of cDNA microarray. Parts of isolated rat livers were used to observe morphology of hepatocytes by light microscopy and electron microscopy. Others were snap frozen in liquid nitrogen and then stored at -80 °C until RNA extraction.
Total RNA was prepared with single-step method by acid guanidinium isothiocyanate-phenol-chloroform according to Chomczynski and Sacchi[8]. In brief, 0.5 g liver was homogenized with 2.5 mL solution D, then 0.25 μL 0.2 mol/L NaAc/0.1 mmol/L ATA, 2.75 μL phenol, 0.55 mL chloroform/isoamyl alcohol (24:1) were added and shaken in order. It was cooled on ice for 15 min and centrifuged at 10000 g at 4 °C for 20 min. The supernatant was removed put into new tubes and 2.75 mL isopropanol was added to precipitate at -20 °C. It was then centrifuged at 10000 g at 4 °C for 20 min. The supernatant was discarded and the pellet was dissolved in RNA denature buffer, then 15 μL 3 mol/LNaAc and 330 μL 1000 μL/L ethanol were added to precipitate at -20 °C for 1 h. Contaminated chromosomal DNA was digested with DNAse I (Promega).
For reverse transcription, RNA (in 1.0 μL DEPC treated water) was mixed with 2.0 μL AP1 (Beckman) and incubated at 70 °C for 5 min, then cooled on ice immediately. The solution was mixed with 8.8 μL DEPC treated water, 4.0 μL 5 × reverse transcript buffer, 2.0 μL dNTP (250 mmol/L), 2.0 μL DTT (100 mmol/L) and 0.2 μL MMLV reverse transcriptase (200 U/μL, GIBCO). Reverse transcription was performed at 42 °C for 5 min, then at 50 °C for 50 min, and finally at 70 °C for 15 min.
For amplification of cDNA, AP1 was chosen as anchor 3’ primer and ARP1-ARP4 were selected as arbitrary 5’ primers. A 2 μL cDNA was mixed with 8.2 μL DEPC treated water, 2.0 μL 10 × PCR buffer (Promega), 2.0 μL MgCl2 (25 mmol/L, Promega), 1.6 μL dNTP (2.5 mmol/L), 2.0 μL ARP (2 mmol/L), 2.0 μL AP (2 mmol/L) and 0.2 μL TaqE (2.0 U/μL, Promega). The PCR amplification program was at 95 °C for 2 min, then 4 cycles at 92 °C for 15 s, at 50 °C for 30 s, and at 72 °C for 2 min, followed by 30 cycles at 92 °C for 15 s, at 60 °C for 30 s, at 72 °C for 2 min, and finally at 72 °C for 7 min.
A 7 μL PCR mixture was redissolved in 4 μL loading dye, heated at 95 °C for 2 min, then run on 80 g/L polyacrylamide-urea gels at 9 mA. DNA fragments on gels were displayed by silver stain method. Bands representing cDNA, which appeared to be differentially expressed, were excised and reamplified under the same conditions as above except the primers. In this procedure, T7 promoter and M13 reverse (-48) were used.
DNA sequence analysis was carried out with dye-terminator method in ABI DNA sequencer. DNA sequences were identified by comparison to those in GenBank BLASTn on Internet.
cDNA probes were prepared through reverse transcription and then purified. The probes from normal rats were labeled with Cy3-dUTP, and the probes from La (NO3) 3-treated rats were labeled with Cy5-dUTP. The chips were scanned using ScanArray3000 laser scanner (General Scanning, Inc) at two wavelengths to detect emissions from both Cy3 and Cy5. The acquired images were analysed using ImaGene3.0 software (BioDiscovery, Inc.). The intensity of each spot at the two wavelengths represented the quantity of Cy3-dUTP and Cy5-dUTP, respectively. Each ratio of Cy3 and Cy5 was computed. Overall intensities were normalized by a coefficient according to the ratios of the located 40 housekeeping genes. To minimize artifacts arising from low expression values, only the genes with raw intensity values larger than 800 counts for one or both of Cy3 and Cy5 were chosen for differential analysis.
La (NO3) 3 had no significant effects on histopathology by light microscopy. Swollen or cavitated mitochondria were observed in hepatocytes by electron microscopy (Figure 1).
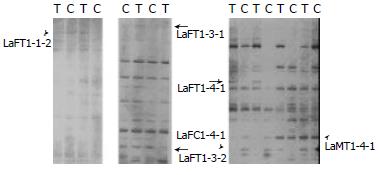
Six differentially expressed bands in two pairs of rats, were obtained (Figure 2). Among them, five were up regulated and one was down regulated in treated rats. Two bands were sequenced. The results were as follows. LaFT1-4-1: The expression level of EST in treated rats was higher than that in control. Its length was about 160 bp (Figure 3A). EST was compared with known ESTs in GeneBank BLASTn. The homology was low (≤ 15%). LaFT1-4-1 was novel. Its sequence was as follow: 5’-AGCGGATAACAATTTCACACAGGA GTAGCAGACCCTGCCCCCAGGAAA TAACACACACTAAACTCTCAAAAA AAAAAAGCCCTATAGTGAGTCGTA TTACACCCTATAGTGAGTCGTATT AGCGGATAACAATTTCACACAGGA CGCCCTATAGTGAGTA-3’. LaMT1-4-1: The expression level of EST in treated rats was higher than that in control. Its length was about 144 bp (Figure 3B). The EST was compared with known ESTs in GenBank BLASTn and it shared 100% sequence homology with AU080263 Sugano mouse brain mncb Mus musculus cDNA clone MNCb-5435 5’. Its sequence was as follow: 5’-ACTCAAAGGCGGTAATACGGTTAT CCACAGAATCAGGGGATAACGCAG GAAAGAACATGTGAGCAAAAGGCC AGCAAAAGGCCAGGAACCGTAAAA AGGCCGCGTTGCTGGCGTTTTTCC ATAGGCTCCGCCCCCCTGACGAGC-3’.
HGEC-40D expression profile microarray, which consisted of 4096 human cDNAs containing 60 control genes and 4036 target genes, was provided by United Gene Holdings, Ltd. For every gene, there were two parallels in microarray. Target genes were divided into 15 types: oncogenes and tumor suppressor genes, ionic passage and transportation protein genes, cell cycle protein genes, stress reaction protein genes, cell skeleton and movement protein genes, genes related to cell apoptosis, DNA synthesis, repair and recombination genes, DNA binding, transcription and transcription factor genes, cell receptor genes, immunity related genes, cell signal and transmission genes, metabolism related genes, protein translation and synthesis genes, growth related genes, and others.
In order to monitor the preparation and hybridization of DNA microarray, positive and negative controls were arranged. Forty housekeeping genes were used as positive controls. 821 gene (8 spots) and 1 × spot solution (8 spots) were used as negative control spots. Positive control spots showed high intensity of signals and negative control spots showed low intensity, which proved the reliability of the data.
Cy3 fluorescent signal (labeled control) and Cy5 fluorescent signal (labeled treated group) were represented with red and green respectively. For overlying two signals of one spot, the spot showed green if the intensity of Cy3 signal was stronger (indicating down-regulation tendency), the spot showed red if the intensity of Cy5 signal was stronger (indicating up-regulation tendency), the spot showed yellow if the intensities of Cy3 and Cy5 signals were similar. In this study, the result of microarray is shown in Figure 4.
We screened out 136 differentially expressed genes according to the following rules. The ratio of Cy3 and Cy5 signal was larger than 2 or smaller than 0.5, the raw intensity value of one or both of Cy3 and Cy5 was larger than 800, the trend of up-regulation or down-regulation in parallels was the same. Of the 136 genes with altered expression, 131 genes had an elevated expression and 5 genes had a reduced expression. Most of the differentially expressed genes were cell signal and transmission genes, genes related to metabolism, protein translation and synthesis genes. Five down-regulated and 19 up-regulated genes with their differential expression levels more than 4-fold and having similar trends, are described in Table 1.
| Trend | GenBank-ID | Definition | Average ratio |
| up | ab023148 | Homo sapiens mRNA for KIAA0931 protein, partial cds. | 23.4 |
| hsu38545 | Human ARF-activated phosphatidylcholine-specific phospholipase D1a (hPLD1) mRNA, complete cds. | 10.1 | |
| hstrke | H.sapiens TRK E mRNA. | 8.7 | |
| humrp17a | Human ribosomal protein L7a (surf 3) large subunit mRNA, complete cds. | 7.8 | |
| humcda24a | Homo sapiens CD24 signal transducer mRNA, complete cds. | 6.9 | |
| hsu76111 | Human translation repressor NAT1 mRNA, complete cds. | 6.4 | |
| af125042 | Homo sapiens bisphosphate 3’-nucleotidase mRNA, complete cds. | 6.1 | |
| hsu43701 | Human ribosomal protein L23a mRNA, complete cds. | 6.0 | |
| hsu85946 | Homo sapiens brain secretory protein hSec10p (HSEC10) mRNA, complete cds. | 5.7 | |
| ae000136 | Escherichia coli K-12 MG1655 section 26 of 400 of the complete genome. | 5.6 | |
| af077951 | Homo sapiens protein inhibitor of activated STAT protein PIAS1 mRNA, complete cds. | 5.3 | |
| af044671 | Homo sapiens MM46 mRNA, complete cds. | 5.1 | |
| hsu35048 | Human TSC-22 protein mRNA, complete cds. | 5.1 | |
| af068302 | Homo sapiens choline/ethanolamine phosphotransferase (CEPT1) mRNA, complete cds. | 5.1 | |
| hsy17392 | Homo sapiens mRNA for prefoldin subunit 1. | 5.0 | |
| af131820 | Homo sapiens clone 25077 mRNA sequence, complete cds. | 4.8 | |
| af094481 | Homo sapiens trinucleotide repeat DNA binding protein p20-CGGBP (CGGBP) gene, complete cds. | 4.8 | |
| ab020636 | Homo sapiens mRNA for KIAA0829 protein, partial cds. | 4.4 | |
| ab020697 | Homo sapiens mRNA for KIAA0890 protein, complete cds. | 4.4 | |
| down | ab011098 | Homo sapiens mRNA for KIAA0526 protein, complete cds. | 0.466 |
| hsifi56r | Human mRNA for 56-ku protein induced by interferon. | 0.437 | |
| hssgk | Homo sapiens sgk gene. | 0.405 | |
| hsu87967 | Human ATP diphosphohydrolase mRNA, complete cds. | 0.364 | |
| humkuant | Human Ku autoimmune antigen gene, complete cds. | 0.352 |
Studies on toxicities of RE elements by different intake pathways showed that the liver was the target organ of RE element toxicity[9-15], and 20 mg/kg was believed to be the effective dose in oral administration[15]. So, rats treated with La (NO3) 3 at a dose of 20 mg/kg by gavage were chosen in our study.
mRNA differential display could provide a unique and powerful experimental system to study differential gene expression. In this study, six differentially expressed tags were obtained. Of the 2 bands sequenced, one (LaFT1-4-1) was novel.
The technique of DNA microarray could allow expression monitoring of thousands of genes in parallel[16]. This technique promotes the identification of differentially expressed genes and investigation of the function of genes. In this study, human cDNA microarray was used. The genomes of human and rodent were very similar[17,18], the homology of human genes and rat genes was over 70%. So it is of significance to analyse rat samples by a HGEC-40D expression profile microarray. One hundred and thirty-six differentially expressed genes were found by DNA microarray, which included mainly cell signal and transmission genes, genes related to metabolism, protein translation and synthesis genes. These genes accounted for 15%, 14% and 13%, respectively. Analysis on capital genes was described as follows.
Some genes encoding mitochondrial proteins were up regulated. The levels of homo sapiens NADH-ubiquinone oxidoreductase B22 subunit mRNA, ND2 gene, NDUFA5 gene and NDUFS8 gene were elevated 2.543, 3.540, 3.645 and 3.767 fold respectively, which were possibly associated with the damage of mitochondria.
Twelve differentially expressed KIAA genes were found in the study. Eleven were up regulated and one was down regulated. The expression level of KIAA0931 was elevated 23.4-fold. A family of KIAA includes new genes published by Kazusa DNA Research Institute in Japan. The functions of KIAA genes were supposed to be related to cell signal transmission, cell structure and cell movement based on the sequence studies[19-21].
Phosphatidylcholine-specific phospholipase D1a (hPLD), is an important member of cell signal transducers. Active hPLD could produce biological effects by the following mechanisms. Phosphatidic acid (PA), diacyl-glycerol (DG) and lysophosphatidic acid (LPA), which are the direct and indirect products of hPLD, are important second messengers. Hydrolysis of phosphatidyl choline (PC) by hPLD could change local components of the membranes, consequently the characteristics of the membranes[22]. It has been revealed that the process of hydrolysis of PC is related to many physiological activities including metabolism, cell cleavage, secretion, immunity, inflammatory reaction[23]. In this study the expression level of hPLD gene was elevated by 10.1-fold, suggesting that activated hPLD which affects cell signal transmission is an important pathway of La (NO3) 3 toxicity.
Signal transducer and activator of transcription (STAT) proteins play an important role in cell proliferation and differentiation induced by cytokines. STAT1 protein is believed to be the transcription factor of IFN reactivity. In addition, the mice with Stat1 gene picked out were found to be lacking of congenital immunity to viruses and bacteria[24]. Protein inhibitor of activated STAT1 (PIAS1) could depress the activity of STAT1 by blocking STAT1 binding to specific DNA[25]. PIAS1 gene was up regulated 5.3-fold in the study. It was suggested that La (NO3) 3 might affect congenital immunity and the reactivity to IFN.
There were 7 differentially expressed genes related to immunity, of which 6 genes were up regulated and 1 was down regulated. In the up-regulated genes, H12.3 protein gene could regulate lymphocyte proliferation[26]. CD9 antigen gene [27], b-globulin gene, immuno-globulin light chain gene, CD24 signal transducer gene were related to humoral immunity. Calnexin protein gene played a role in cellular immunity and humoral immunity[28]. The results suggested that La (NO3) 3 had effects on both cellular immunity and humoral immunity. This was in good agreement with the results obtained by other researchers[29-31].
In conclusion, the expression profiles of certain genes of La (NO3) 3 treated rats differed markedly from those of control rats. This is consistent with the wide biological effect spectrum of RE elements. Multiple genes may join together to play a same role. Further analysis of the differentially expressed genes would be helpful for understanding the wide biological effect spectrum of RE elements. It would be interesting to explore further if the differentially expressed genes could be used as more sensitive biomarkers.
| 1. | Xu HE, Gao GH, Jia FL, Wangh XY, Xie Q, Liu HS, Wang NF. Effect of mixed rare earth Changle on the micronuclei forma-tion of blood lymphocytes in rats. Zhonghua Yufang Yixue Zazhi. 2000;34:5-7. |
| 2. | Nakamura Y, Tsumura Y, Tonogai Y, Shibata T, Ito Y. Differences in behavior among the chlorides of seven rare earth elements administered intravenously to rats. Fundam Appl Toxicol. 1997;37:106-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 91] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 3. | Tuchweber B, Trost R, Salas M, Sieck W. Effect of praseodymium nitrate on hepatocytes and Kupffer cells in the rat. Can J Physiol Pharmacol. 1976;54:898-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 12] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 4. | Salas M, Tuchweber B, Kovacs K, Garg BD. Effect of cerium on the rat liver: an ultrastructural and biochemical study. Beitr Pathol. 1976;157:23-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 5. | Li XQ, Jiang JJ, Yang L, Liu M, Yang LY. Effect of low level mixed rare earth Changle on learning and memory, locomotor activity and NMDA-and M-receptors activity and morphol-ogy of divisions of hippocampal cortex in rats. Zhonghua Yufang Yixue Zazhi. 2000;34:20-23. |
| 6. | Liu JM, Chen D, Wang XM, Nie YX, Li Y, Li J. Long-term effect of Changle and La (NO3) 3 on level of IL-2 and g-IFN of splenic lymphocytes in rats. Zhonghua Yufang Yixue Zazhi. 2000;34:49-51. |
| 7. | Guo XB, Gao ZH, Luo LZ, Yao BY. Effects of rare earth com-pounds on metabolic cooperation between Chinese hamster V79 cells. Zhonghua Yufang Yixue Zazhi. 2000;34:17-19. |
| 8. | Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40518] [Cited by in RCA: 39212] [Article Influence: 1005.4] [Reference Citation Analysis (0)] |
| 9. | Godin DV, Frohlich J. Erythrocyte alterations in praseodymium-induced lecithin: cholesterol acyltransferase (LCAT) deficiency in the rat: comparison with familial LCAT deficiency in man. Res Commun Chem Pathol Pharmacol. 1981;31:555-566. [PubMed] |
| 10. | Langer GA, Frank JS. Lanthanum in heart cell culture. Effect on calcium exchange correlated with its localization. J Cell Biol. 1972;54:441-455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 201] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 11. | Arvela P, Kraul H, Stenbäck F, Pelkonen O. The cerium-induced liver injury and oxidative drug metabolism in DBA/2 and C57BL/6 mice. Toxicology. 1991;69:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 12. | Spencer A, Wilson S, Harpur E. Gadolinium chloride toxicity in the mouse. Hum Exp Toxicol. 1998;17:633-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 13. | Hirano S, Kodama N, Shibata K, Suzuki KT. Metabolism and toxicity of intravenously injected yttrium chloride in rats. Toxicol Appl Pharmacol. 1993;121:224-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 31] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 14. | Shinohara A, Chiba M, Inaba Y. Distribution of terbium and increase of calcium concentration in the organs of mice i.v.-administered with terbium chloride. Biomed Environ Sci. 1997;10:73-84. [PubMed] |
| 15. | Chen AJ, Chen D, Liu Y, Liu P, Wang XM, Nie YX, Wang X, Sun SY. The long term effect of low dose of Changle on thestructure and function of liver in rats. Zhonghua Yufang YixueZazhi. 2000;34:46-48. |
| 16. | Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6477] [Cited by in RCA: 5132] [Article Influence: 165.5] [Reference Citation Analysis (0)] |
| 17. | Pennisi E. Genomics. Sequence tells mouse, human genome secrets. Science. 2002;298:1863-1865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 18. | Pennisi E. Genomics. Charting a genome's hills and valleys. Science. 2002;296:1601-1603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 19. | Nagase T, Ishikawa K, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5:31-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 150] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 20. | Nagase T, Ishikawa K, Suyama M, Kikuno R, Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. XII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1998;5:355-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 182] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 21. | Nagase T, Ishikawa K, Suyama M, Kikuno R, Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. XIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1999;6:63-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 105] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 22. | Exton JH. Phospholipase D: enzymology, mechanisms of regulation, and function. Physiol Rev. 1997;77:303-320. [PubMed] |
| 23. | Morris AJ, Engebrecht J, Frohman MA. Structure and regulation of phospholipase D. Trends Pharmacol Sci. 1996;17:182-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 137] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 24. | Coffman RL, Lebman DA, Rothman P. Mechanism and regulation of immunoglobulin isotype switching. Adv Immunol. 1993;54:229-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 386] [Cited by in RCA: 403] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 25. | Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, Shuai K. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci USA. 1998;95:10626-10631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 560] [Cited by in RCA: 566] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 26. | Guillemot F, Billault A, Auffray C. Physical linkage of a guanine nucleotide-binding protein-related gene to the chicken major histocompatibility complex. Proc Natl Acad Sci USA. 1989;86:4594-4598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 84] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Boucheix C, Benoit P, Frachet P, Billard M, Worthington RE, Gagnon J, Uzan G. Molecular cloning of the CD9 antigen. A new family of cell surface proteins. J Biol Chem. 1991;266:117-122. [PubMed] |
| 28. | Hochstenbach F, David V, Watkins S, Brenner MB. Endoplasmic reticulum resident protein of 90 kilodaltons associates with the T- and B-cell antigen receptors and major histocompatibility complex antigens during their assembly. Proc Natl Acad Sci USA. 1992;89:4734-4738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 202] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 29. | Wang YZ, Li ZX, Li F, Li M, Shi Y. Effects of RE elements on workers’ immune function. Zhiye Yixue. 1995;22:7-8. |
| 30. | Zhang ZS, Xue B, Chen XA. Effects of RE nitrate on the func-tion of T- lymphocytes, B- lymphocytes and macrophage. Weisheng Dulixue Zazhi. 1993;7:157-158. |
| 31. | Wei XT, Ma N, Lei ZM, Xue B, Xu HE. Effects of La (NO3) 3 on the immune function and cell apoptosis of splenic lympho-cytes and thymocytes. Zhonghua Yufang Yixue Zazhi. 2000;34:42-45. |
Edited by Zhu LH and Wang XL Proofread by Xu FM