©The Author(s) 2026.
World J Gastroenterol. Feb 21, 2026; 32(7): 113973
Published online Feb 21, 2026. doi: 10.3748/wjg.v32.i7.113973
Published online Feb 21, 2026. doi: 10.3748/wjg.v32.i7.113973
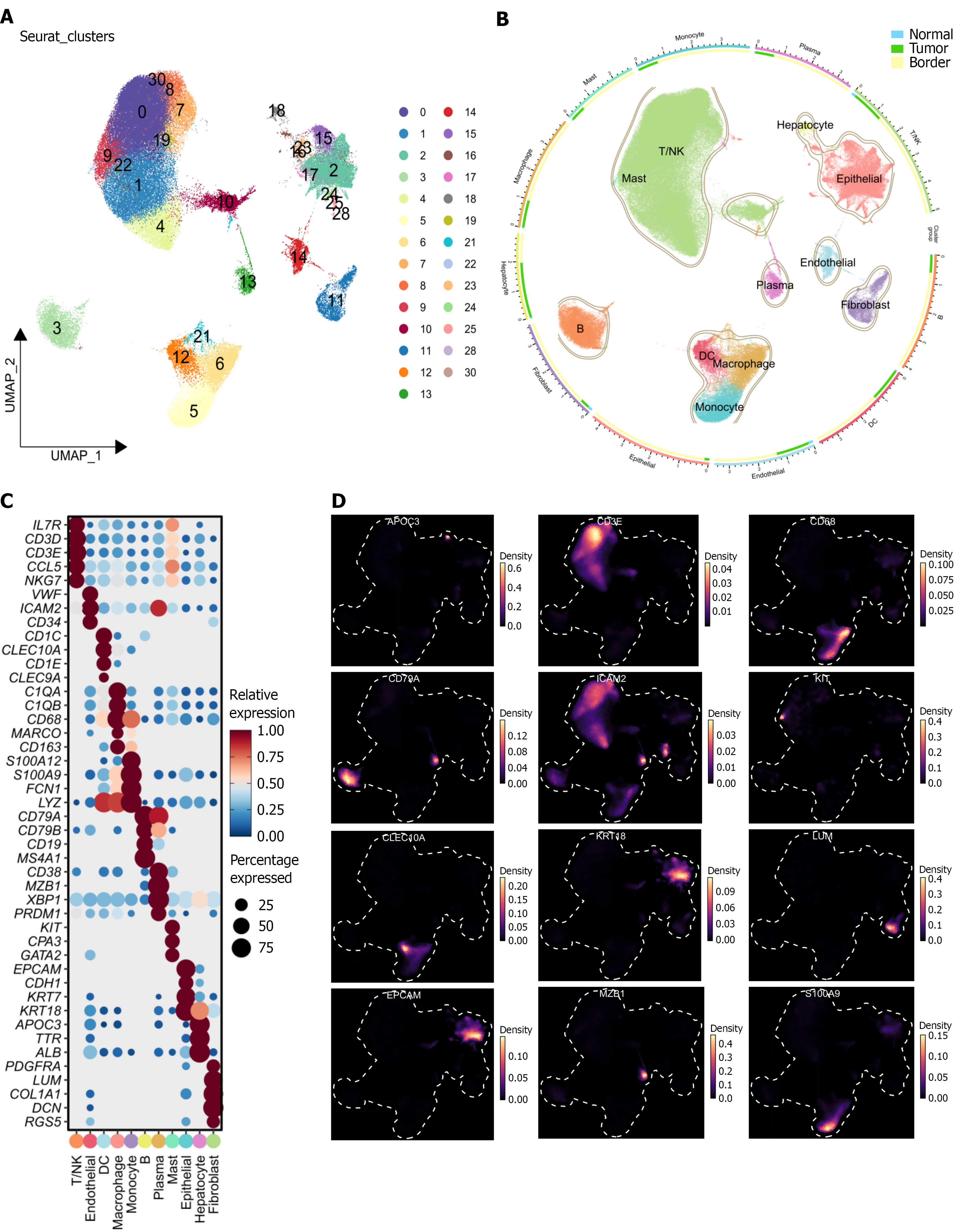
Figure 1 Single-cell landscape and cell-type annotation of intrahepatic cholangiocarcinoma.
A: Uniform manifold approximation and projection (UMAP) projection identifying 31 transcriptionally distinct clusters; B: Cell-type annotation based on canonical marker gene expression, revealing 11 major populations; C: Dot-plot visualization of representative marker genes used for each annotated cell type and their relative expression level; D: UMAP feature plots showing spatial expression patterns of selected canonical markers. UMAP: Uniform manifold approximation and projection; NK: Natural killer; DC: Dendritic cell.
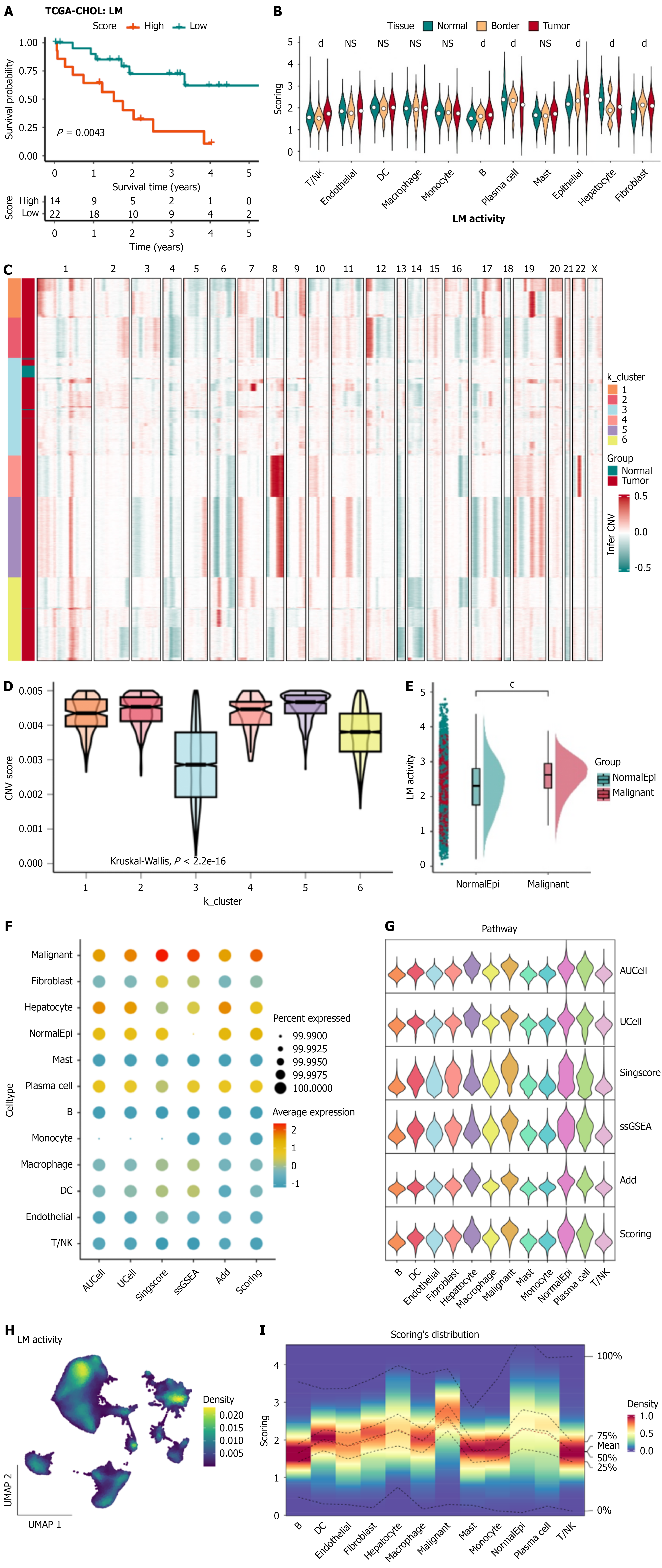
Figure 2 Evaluation of lactate metabolism activity in intrahepatic cholangiocarcinoma.
A: Kaplan-Meier analysis of the Cancer Genome Atlas-cholangiocarcinoma cohort showing that patients with high lactate metabolism (LM) activity exhibited significantly reduced overall survival compared to the low LM group (P = 0.0043); B: Violin plots comparing LM activity across major cell types within the intrahepatic cholangiocarcinoma (ICC) microenvironment; C: Heatmap of infer copy number variation (CNV) analysis identifying six epithelial clusters based on chromosomal CNV profiles; D: Boxplots illustrating significantly higher CNV scores in malignant clusters (K1, K2, K4, K5, and K6) compared with non-malignant cluster K3 (P < 0.001); E: Comparison of LM activity between normal and malignant epithelial cells, showing markedly elevated LM activity in the malignant population (P < 0.001); F and G: LM activity quantified by five computational algorithms (AUCell, UCell, singscore, single-sample Gene Set Enrichment Analysis, and AddModuleScore), consistently indicating higher LM activity in malignant epithelial cells; H: Uniform manifold approximation and projection visualization displaying the spatial distribution of LM activity scores across the ICC microen
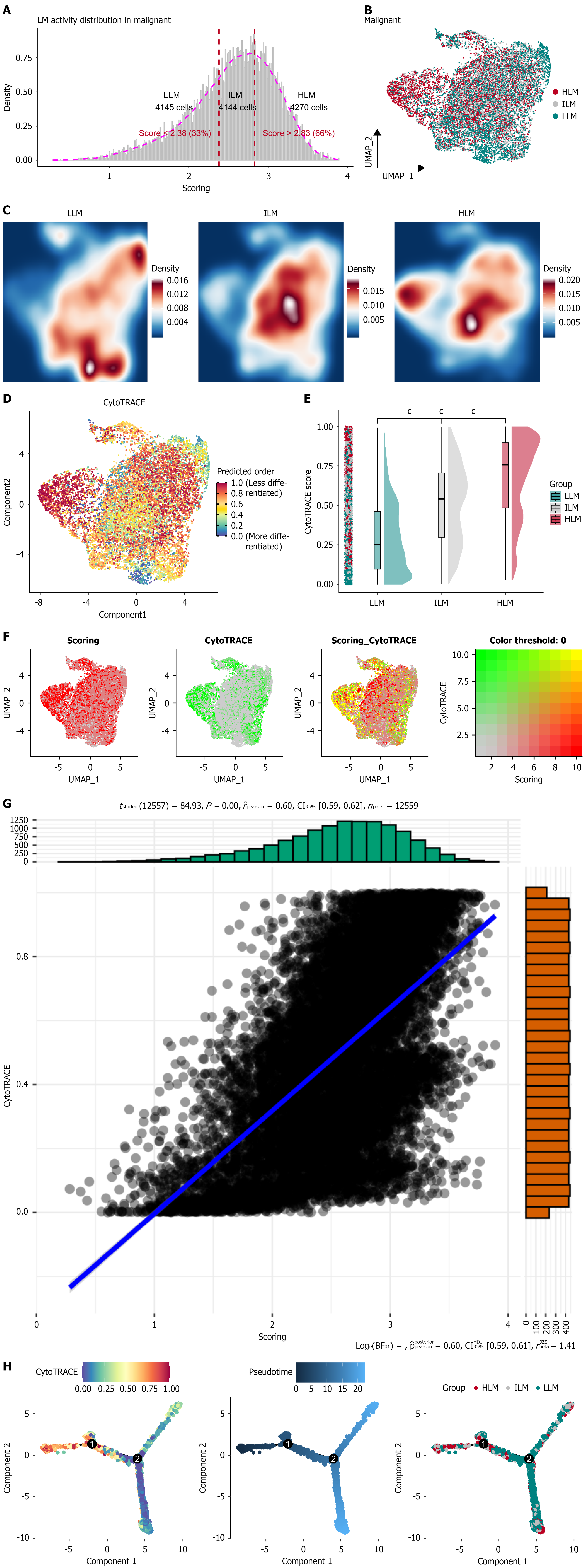
Figure 3 Lactate metabolism heterogeneity intrahepatic cholangiocarcinoma malignant cells.
A: Distribution of lactate metabolism (LM) activity scores in malignant epithelial cells. Cells were stratified into low-LM (LLM) (< 2.38), intermediate-LM (ILM), and high-LM (HLM) (> 2.83) activity groups; B: Spatial distribution of LLM, ILM, and HLM subgroups; C: Density maps illustrating distinct spatial patterns of LM activity; D: Differentiation potential across malignant cells; E: CytoTRACE scores among subgroups; F: The co-expression relationship of LM scoring and CytoTRACE score; G: Correlation plot of LM activity and CytoTRACE scores (r = 0.60, Pearson), showing a strong positive association; H: The development trajectory of malignant epithelial cell subpopulation. cP < 0.001. LM: Lactate metabolism; LLM: Low-lactate metabolism; ILM: Intermediate-lactate metabolism; HLM: High-lactate metabolism; UMAP: Uniform manifold approximation and projection; CI: Confidence interval.
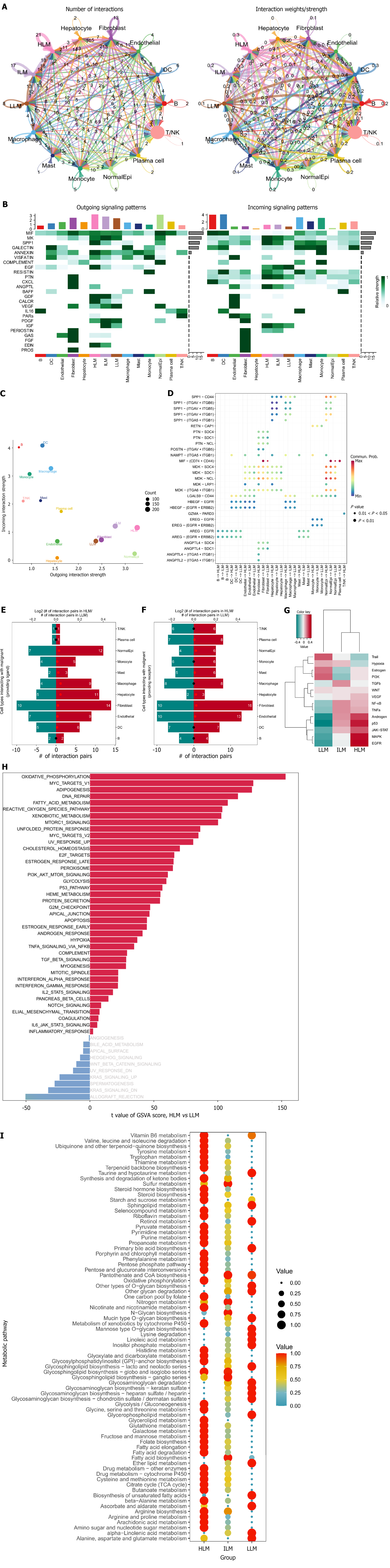
Figure 4 Biological function analysis of lactate metabolism activity subgroups in intrahepatic cholangiocarcinoma.
A: Number and strength of intercellular interactions in intrahepatic cholangiocarcinoma (ICC) tumor microenvironment; B: Outgoing and incoming signaling patterns in ICC microenvironment; C: Scatter plot comparing outgoing vs incoming interaction strength of cell types; D: Ligand-receptor interaction analysis for malignant lactate metabolism (LM) cells; E and F: Interaction pairs of cell types interacting with malignant LM activity cells in providing ligand and receptor; G: PROGENy pathway activity heatmap showing elevated oncogenic signaling in LM activity subgroups; H: GSVA enrichment of hallmark pathways distinguishing high-LM and low-LM; I: Metabolic pathway enrichment analysis in LM activity subgroups. NK: Natural killer; DC: Dendritic cell; LLM: Low-lactate metabolism; ILM: Intermediate-lactate metabolism; HLM: High-lactate metabolism; PI3K: Phosphatidylinositol 3-kinase; TGF: Transforming growth factor; VEGF: Vascular endothelial growth factor; NF-κB: Nuclear factor kappa-B; TNF: Tumor necrosis factor; JAK-STAT: Janus kinase-signal transducer and activator of transcription; MAPK: Mitogen-activated protein kinase; EGFR: Epidermal growth factor receptor.
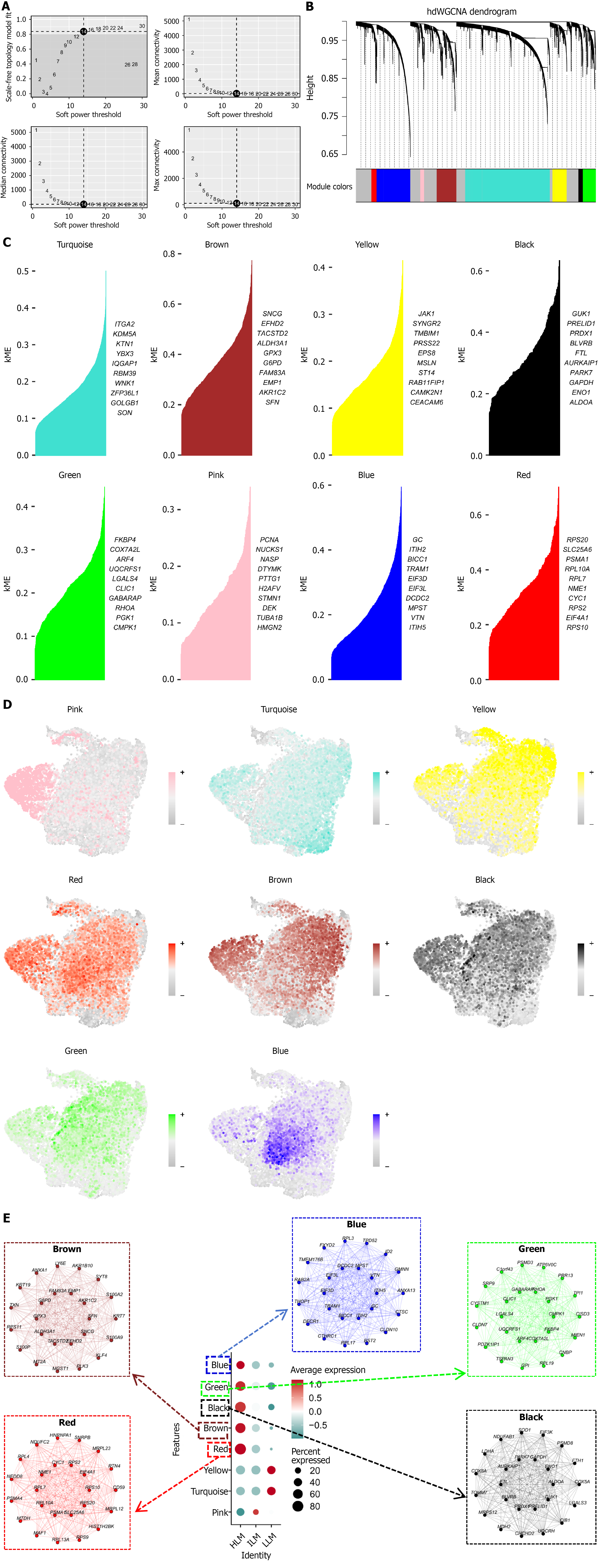
Figure 5 Identification of lactate metabolism-associated gene co-expression modules in malignant intrahepatic cholangiocarcinoma cells.
A: Determination of soft-thresholding power (β = 14) achieving scale-free topology fit ≥ 0.8; B: Gene dendrogram and module color assignment by high-dimensional weighted gene co-expression network analysis; C: Module eigengene correlations and top hub genes per module; D: Uniform manifold approximation and projection visualization of module-specific gene expression across malignant cells; E: Module-trait relationship plot showing relative enrichment of modules across lactate metabolism activity subgroups. hdWGCNA: High-dimensional weighted gene co-expression network analysis; kME: Eigengene connectivity; LLM: Low-lactate metabolism; ILM: Intermediate-lactate metabolism; HLM: High-lactate metabolism.
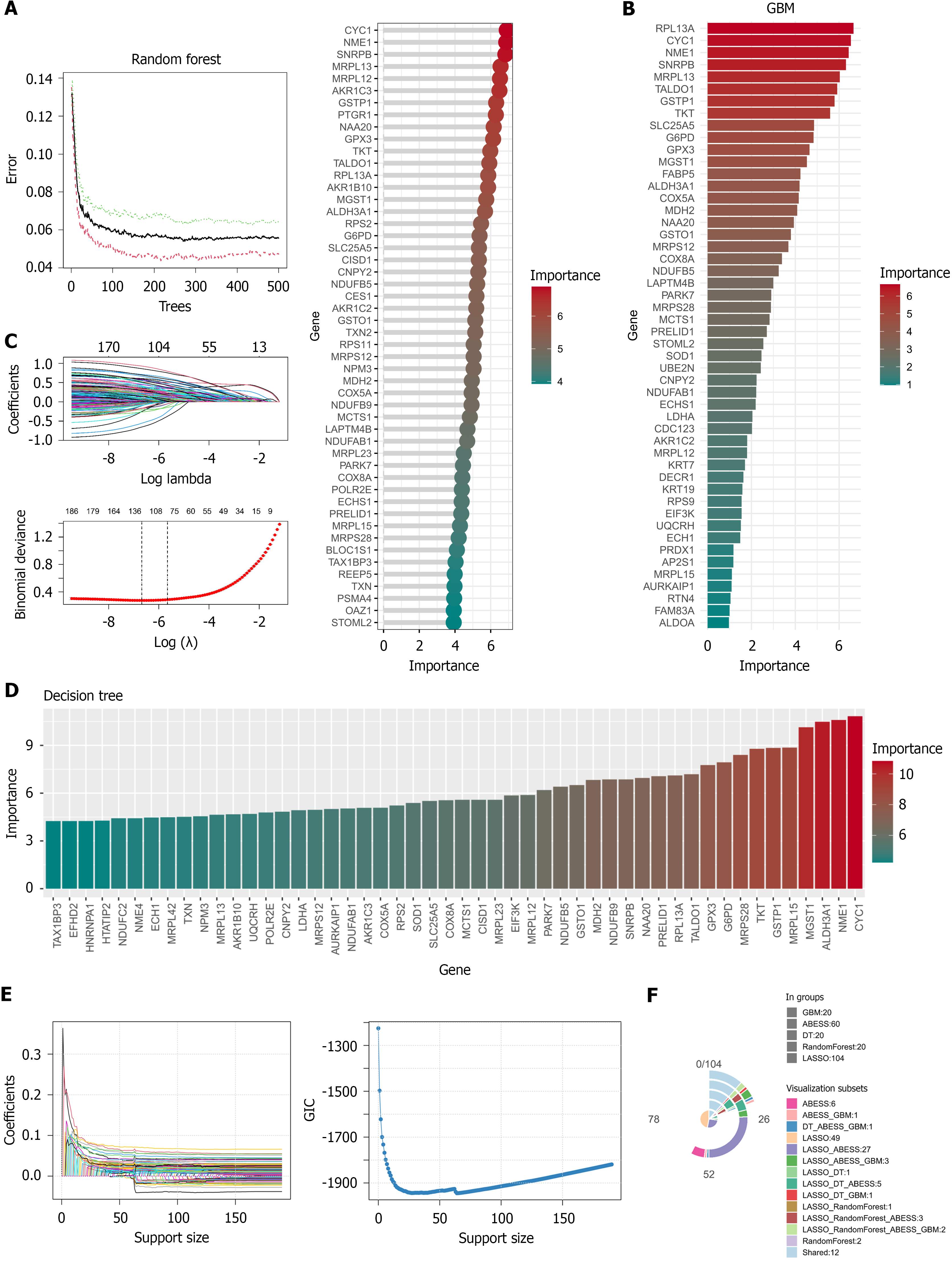
Figure 6 Multi-algorithm identification of lactate metabolism-related feature genes.
A: Importance of signature genes evaluated in random forest; B: Importance of signature genes generated from gradient boosting machine; C: Optimal lambda tuning and cross-validation in least absolute shrinkage and selection operator; D: Importance of signature genes generated from decision tree; E: Candidate optimal signature genes obtained in adaptive best subset selection; F: Intersection diagram summarizing overlapping genes across the five algorithms, identifying 12 shared lactate metabolism-associated genes. GBM: Gradient boosting machine; DT: Decision tree; ABESS: Adaptive best subset selection; LASSO: Least absolute shrinkage and selection operator.
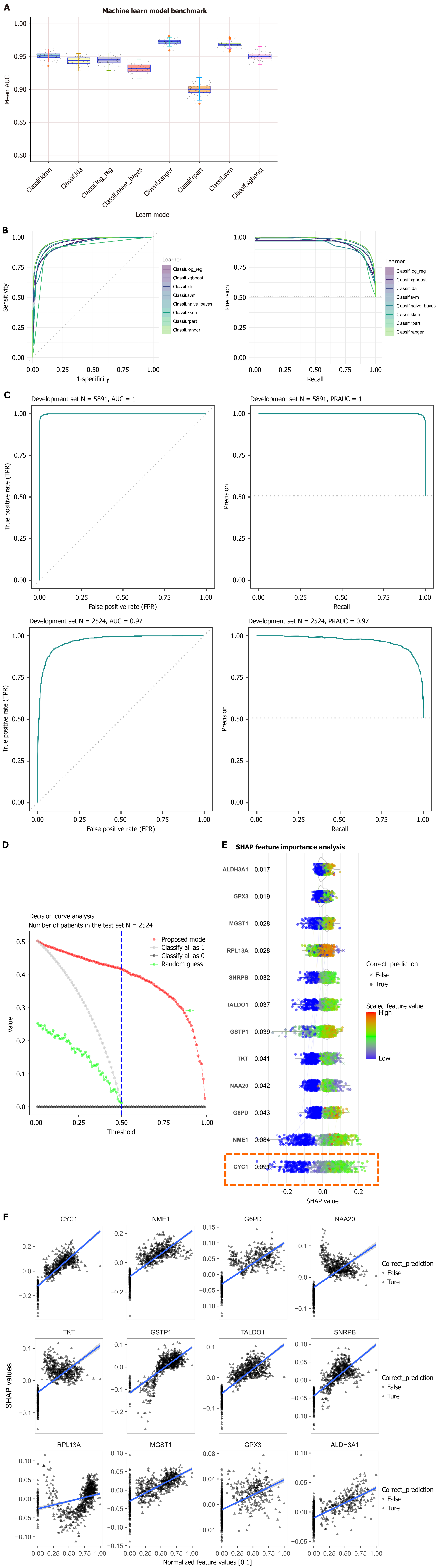
Figure 7 Benchmarking and interpretation of the optimal lactate metabolism-prediction model.
A: Benchmark analysis for model comparison; B: Receiver operating characteristic (ROC) and precision-recall (PR) curves examined the accuracy of different classifiers; C: ROC and PR curves confirming random forest model accuracy [area under the curve (AUC) = 0.97, PRAUC = 0.97]; D: Decision curve analysis for clinical applicability assessment; E: SHapley Additive exPlanation (SHAP) feature importance analysis of 12 lactate metabolism-related genes; F: Correlation between SHAP values and the expression of feature genes. AUC: Area under the curve; PR: Precision-recall; TPR: True positive rate; FPR: False positive rate; SHAP: SHapley Additive exPlanation.
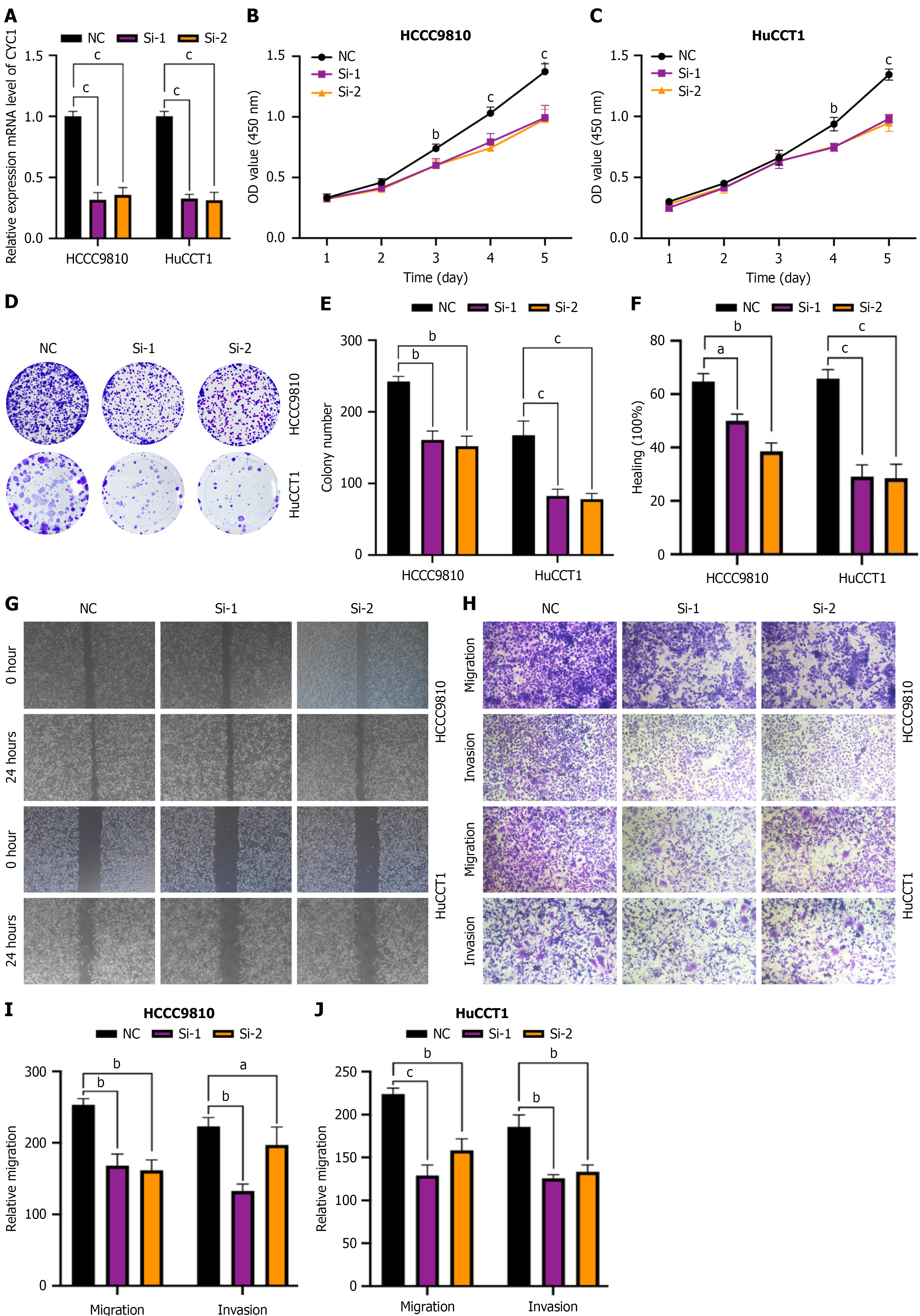
Figure 8 Functional role of CYC1 in intrahepatic cholangiocarcinoma cell lines.
A: Quantitative polymerase chain reaction validation of CYC1 knockdown efficiency in HCCC9810 and HuCCT1 cells; B and C: Cell counting kit-8 assay showing reduced proliferation following CYC1 silencing; D and E: Colony-formation assay demonstrating decreased colony number and size after CYC1 knockdown; F and G: Wound-healing assay showing impaired migratory capacity in CYC1-deficient cells; H-J: Transwell migration and invasion assays confirming that CYC1 depletion significantly suppresses motility in both intrahepatic cho
- Citation: Wu AK, Li JY, Zhang K, Meng M, Wang X, Liu Y, Xie P, Rong WQ, Wu F, Wang HG, Meng X, Wu JX. Lactate metabolism-driven tumor heterogeneity and molecular signatures in intrahepatic cholangiocarcinoma. World J Gastroenterol 2026; 32(7): 113973
- URL: https://www.wjgnet.com/1007-9327/full/v32/i7/113973.htm
- DOI: https://dx.doi.org/10.3748/wjg.v32.i7.113973