©The Author(s) 2026.
World J Gastroenterol. Jan 7, 2026; 32(1): 111428
Published online Jan 7, 2026. doi: 10.3748/wjg.v32.i1.111428
Published online Jan 7, 2026. doi: 10.3748/wjg.v32.i1.111428
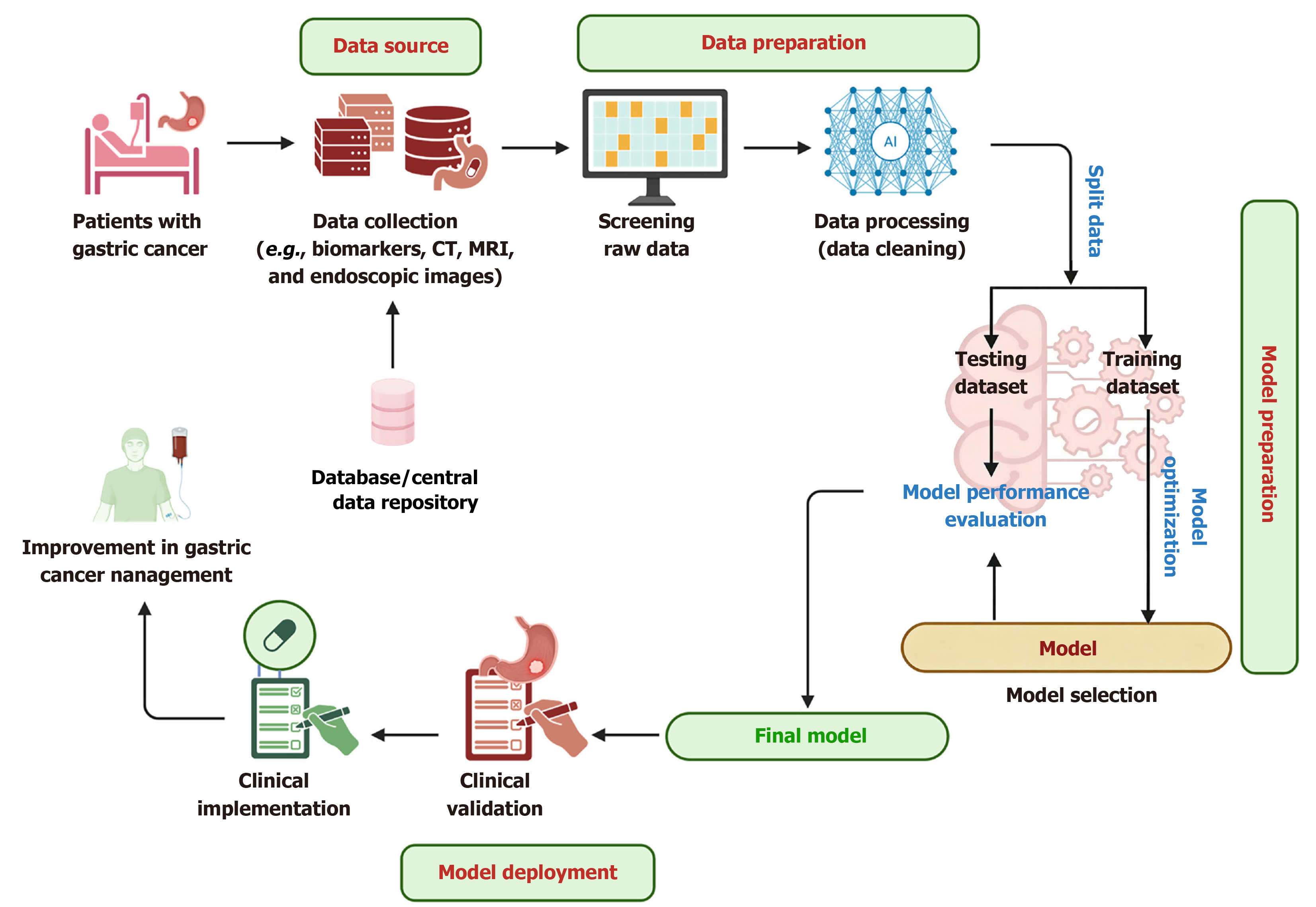
Figure 1 Schematic representation of a general artificial intelligence-based workflow in gastrointestinal cancer research.
AI: Artificial in
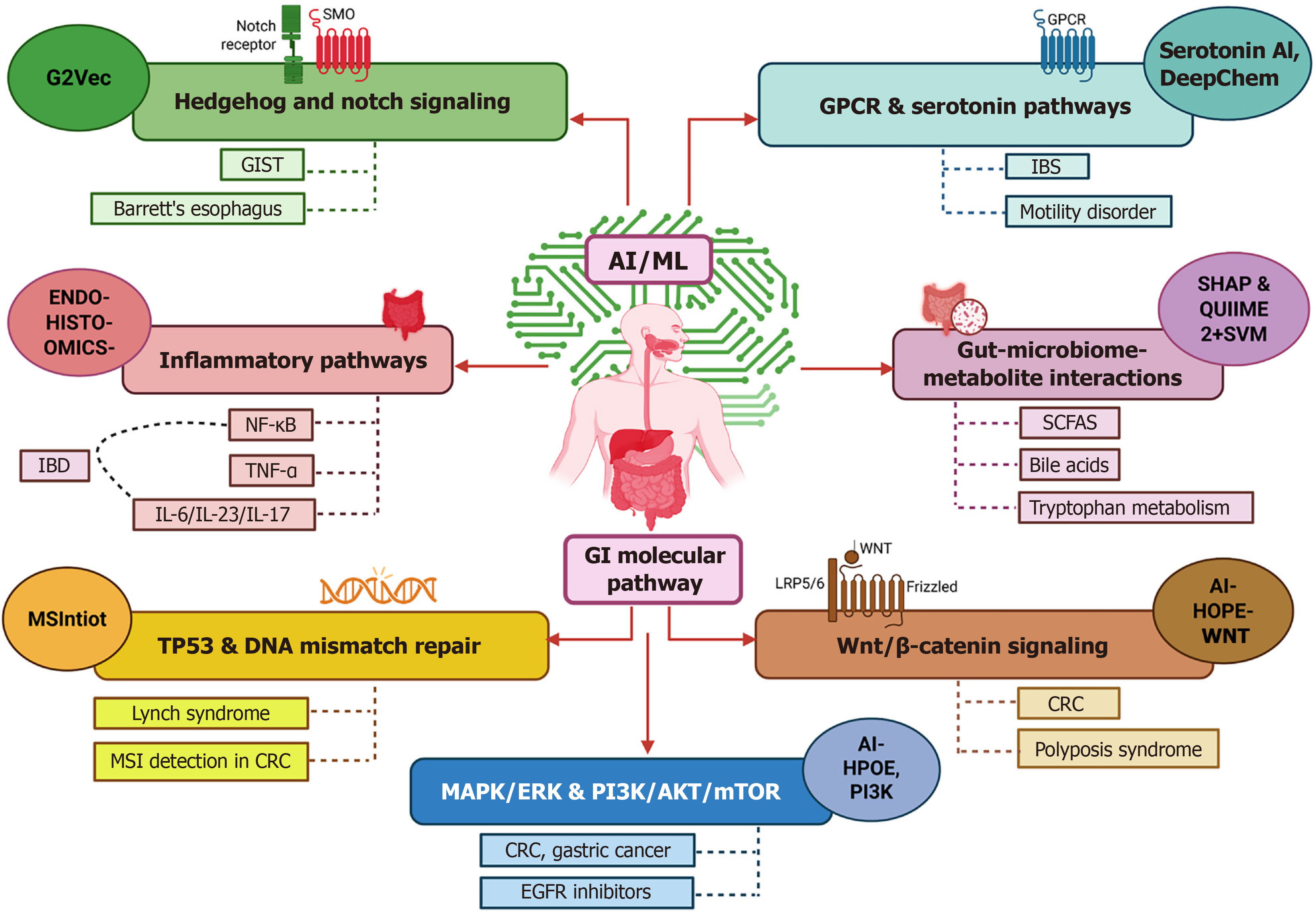
Figure 2 Artificial intelligence-driven analysis of molecular pathways in gastrointestinal cancers and associated disorders.
This figure illustrates how artificial intelligence (AI) helps decode key molecular pathways involved in gastrointestinal diseases. Tools like G2Vec (a graph-based AI) are used for pathway analysis, while Hedgehog and Notch signaling pathways are linked to conditions such as Barrett’s esophagus and gastrointestinal stromal tumors. ENDO-HISTO-OMICS integrates endoscopic, histological, and multi-omics data through AI to enhance diagnosis. Inflammatory pathways like nuclear factor kappa-B, tumor necrosis factor-α, and the interleukin (IL)-6/IL-23/IL-17 are evaluated in inflammatory bowel disease. AI also detects TP53 mutations and DNA mismatch repair defects, which are associated with Lynch syndrome and microsatellite instability in colorectal cancer (CRC). Deep learning tools such as DeepChem are applied to analyze G protein-coupled receptor and serotonin pathways, which play roles in irritable bowel syndrome and gut motility disorders. Models like QUIIME 2, paired with support vector machines, help explore interactions between the gut microbiome and metabolites such as short-chain fatty acids, bile acids, and tryptophan derivatives. AI also informs responses to therapies by analyzing key signaling pathways including Wnt/β-catenin and mitogen-activated protein kinase/extracellular regulated protein kinases/phosphatidylinositol 3-kinase/protein kinase B/mammalian target of rapamycin in CRC and gastric cancers. SMO: Site management organization; GIST: Gastrointestinal stromal tumor; AI: Artificial intelligence; GPCR: G protein-coupled receptor; IBS: Irritable bowel syndrome; ML: Machine learning; IBD: Inflammatory bowel disease; NF-κB: Nuclear factor kappa-B; TNF: Tumor necrosis factor; IL: Interleukin; SHAP: SHapley Additive exPlanations; SVM: Support vector machine; SCFAS: Short chain fatty acid; GI: Gastrointestinal; MSI: Microsatellite Instability; CRC: Colorectal cancer; MAPK: Mitogen-activated protein kinase; ERK: Extracellular regulated protein kinases; PI3K: Phosphatidylinositol 3-kinase; AKT: Protein kinase B; mTOR: Mammalian target of rapamycin; EGFR: Epidermal growth factor receptor.
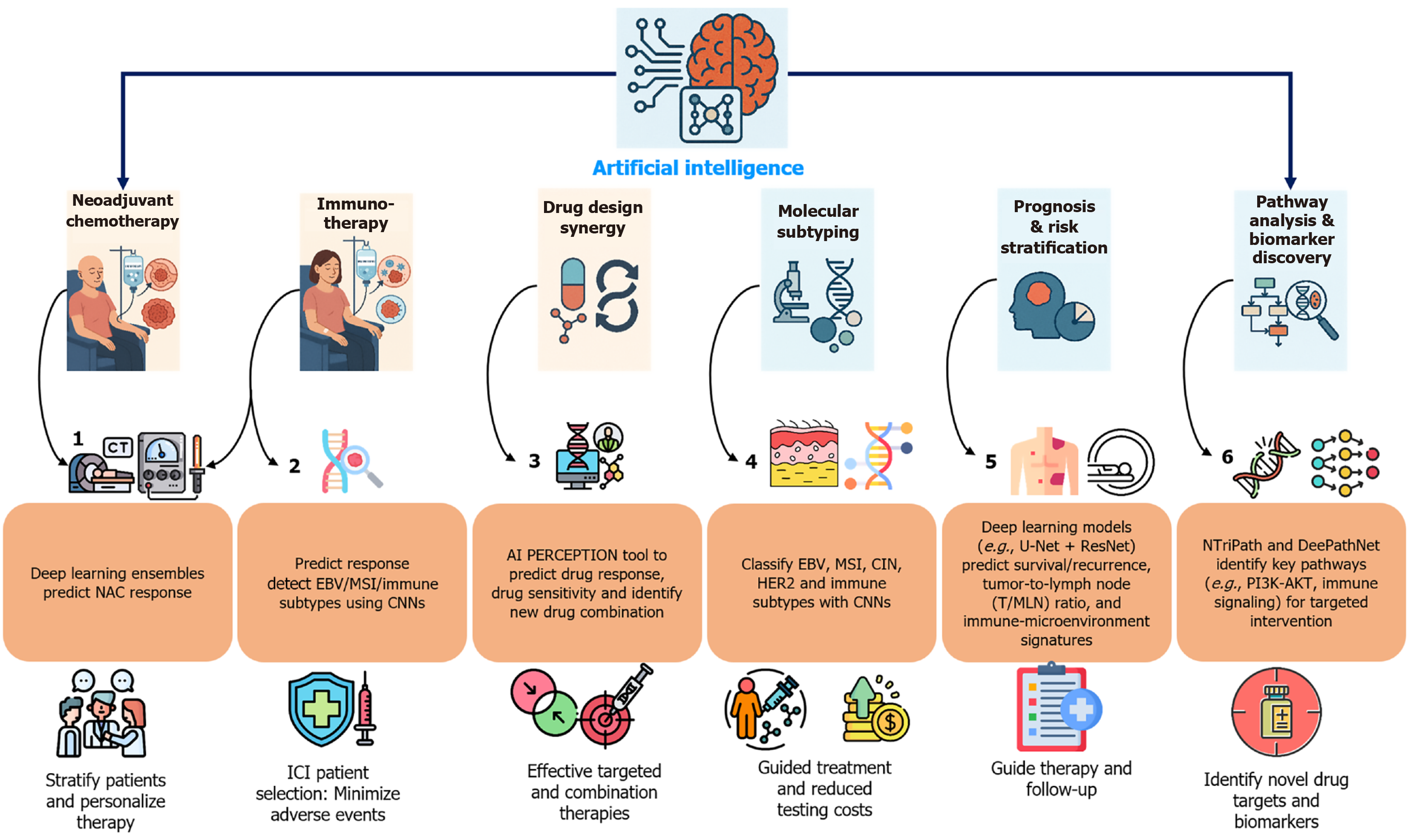
Figure 3 Artificial intelligence applications in gastrointestinal cancer therapeutics.
This schematic illustrates artificial intelligence approaches for neoadjuvant chemotherapy optimization and precision oncology. Panel labels (1-6) denote data sources: (1) Computed tomography (CT) scans + radiomics; (2) Radiomics + gene expression; (3) Multi-omics (genomics of drug sensitivity in cancer/cancer cell line encyclopedia); (4) Histology + genomics; (5) Whole slide imaging, CT, and immunohistochemistry; and (6) Somatic mutations + networks. NAC: Neoadjuvant chemotherapy; EBV: Epstein-Barr virus; MSI: Microsatellite instability; CNN: Convolutional neural network; CIN: Chromosomal instability; T/MLN: Tumor/metastasis lymph node; ICI: Immune checkpoint inhibitor.
- Citation: Suri C, Ratre YK, Pande B, Bhaskar L, Verma HK. Artificial intelligence and machine learning-driven advancements in gastrointestinal cancer: Paving the way for precision medicine. World J Gastroenterol 2026; 32(1): 111428
- URL: https://www.wjgnet.com/1007-9327/full/v32/i1/111428.htm
- DOI: https://dx.doi.org/10.3748/wjg.v32.i1.111428