©The Author(s) 2025.
World J Gastroenterol. Jul 28, 2025; 31(28): 108990
Published online Jul 28, 2025. doi: 10.3748/wjg.v31.i28.108990
Published online Jul 28, 2025. doi: 10.3748/wjg.v31.i28.108990
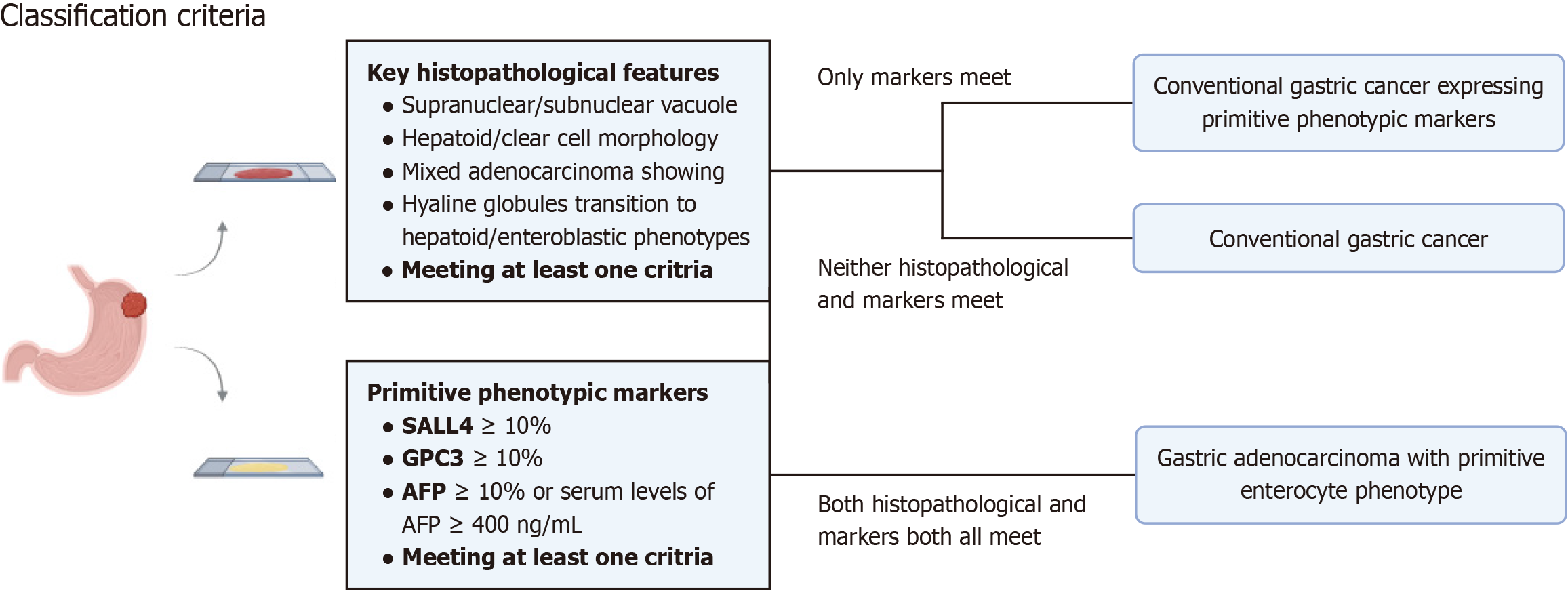
Figure 1 The classification criteria of various histological gastric cancer subtypes.
AFP: Alpha-fetoprotein; SALL4: Spalt-like transcription factor 4; GPC3: Glypican-3.
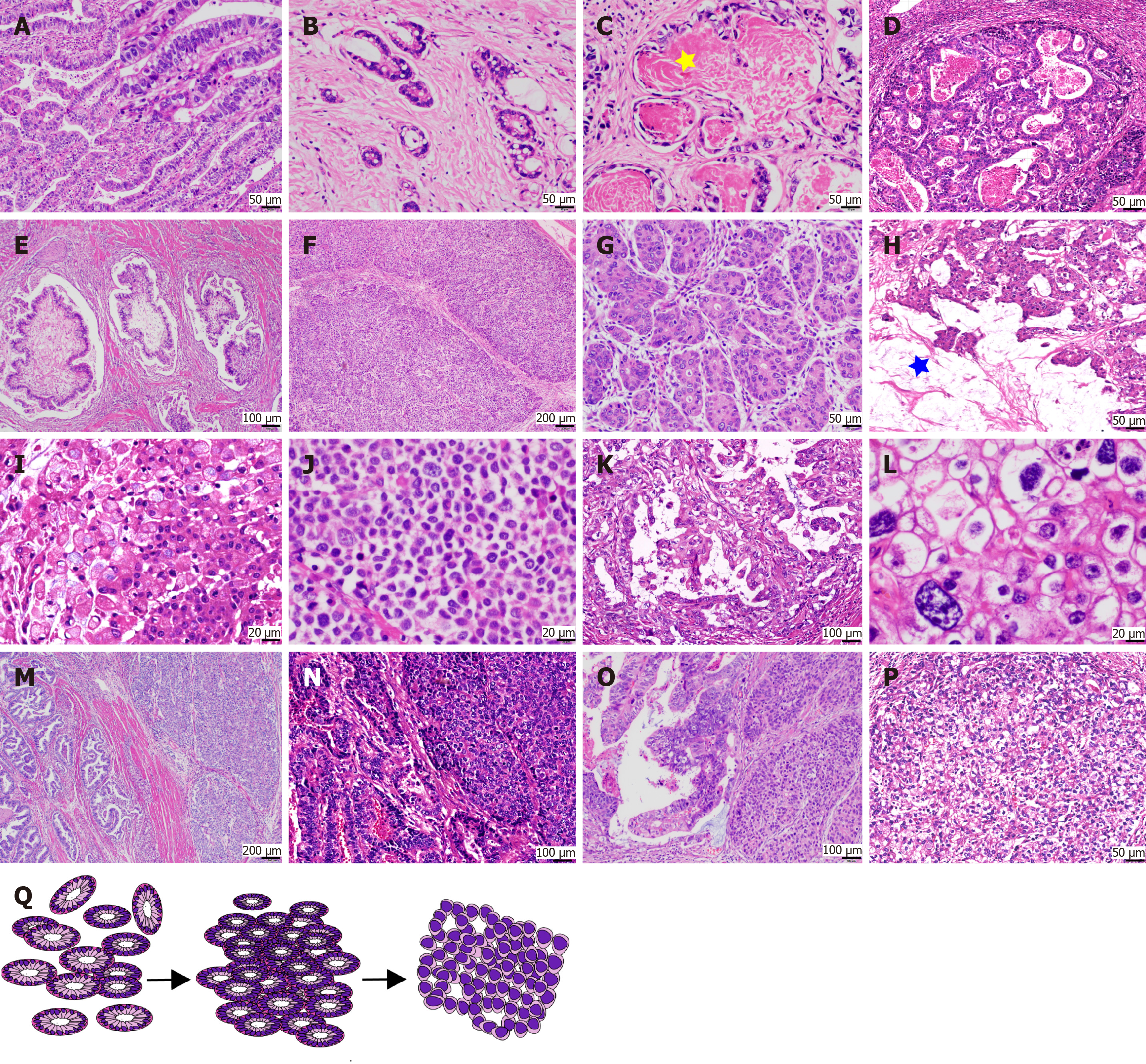
Figure 2 Histopathological diversity of gastric adenocarcinoma with primitive enterocyte phenotype.
A and B: Tubular-papillary subtype of gastric adenocarcinoma with primitive enterocyte phenotype characterized by glandular structures with supranuclear and subnuclear vacuoles, creating a distinctive “piano keyboard-like” architecture; C: Luminal eosinophilic secretions within glandular spaces (asterisks); D and E: Uncommon architectural patterns, including sieve-like/sac-like formations and villous projections with tumor cells exhibiting flattened morphology; F and G: Solid subtype resembling hepatocellular carcinoma, featuring large polygonal tumor cells with eosinophilic or clear cytoplasm arranged in nested sheets; H and I: Intracellular and extracellular mucin deposition (asterisks) observed in select cases; J: Rare poorly cohesive growth pattern akin to gastric poorly cohesive carcinoma; K and L: High-grade cytological atypia, including nuclear pleomorphism and prominent nucleoli; M-O: Transitional zones demonstrating admixture and migratory progression between hepatoid adenocarcinoma and gastric adenocarcinoma with enteroblastic differentiation components; P: Predominance of clear cytoplasmic features over eosinophilic differentiation in solid areas; Q: Schematic representation of phenotypic evolution from tubular-papillary to solid morphology.
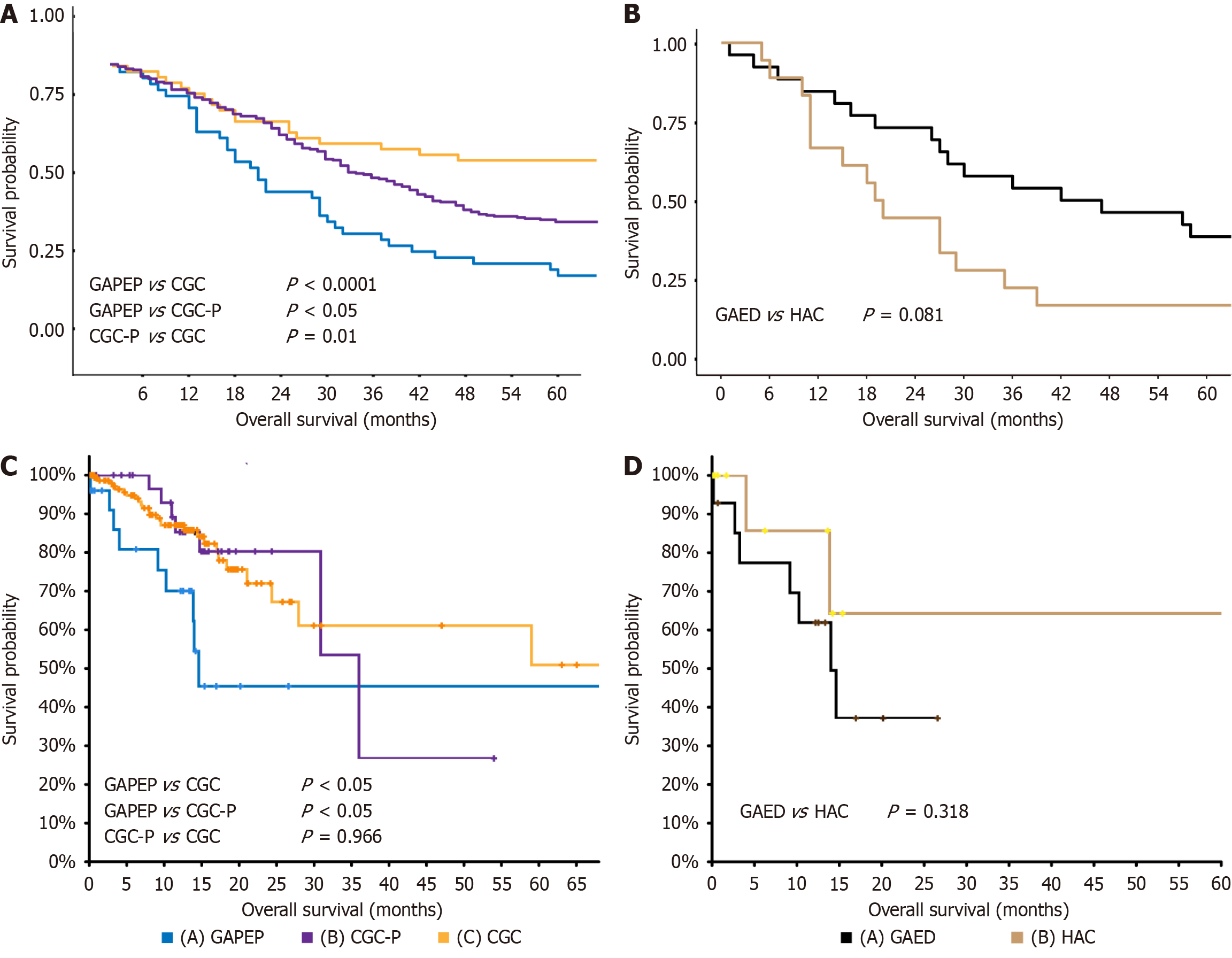
Figure 3 Prognostic significance of gastric adenocarcinoma with primitive enterocyte phenotype compared to other gastric cancer subtypes.
A: Kaplan-Meier survival curves using our institutional cohort demonstrate significantly worse overall survival (OS) in gastric adenocarcinoma with primitive enterocyte phenotype patients vs conventional gastric cancer and conventional gastric cancer expressing primitive phenotypic markers; B: Stratified analysis across cohorts (in-house); C: Kaplan-Meier survival curves using The Cancer Genome Atlas data demonstrate significantly worse OS in gastric adenocarcinoma with primitive enterocyte phenotype patients vs conventional gastric cancer and conventional gastric cancer expressing primitive phenotypic markers; D: Stratified analysis across cohorts (The Cancer Genome Atlas showed no OS difference between hepatoid adenocarcinoma and gastric adenocarcinoma with enteroblastic differentiation (P = 0.081 and P = 0.318, respectively), supporting their shared clinicopathological spectrum. GAPEP: Gastric adenocarcinoma with primitive enterocyte phenotype; CGC: Conventional gastric cancer; CGC-P: CGC: Conventional gastric cancer expressing primitive phenotypic markers; HAC: Hepatoid adenocarcinoma; GAED: Gastric adenocarcinoma with enteroblastic differentiation.
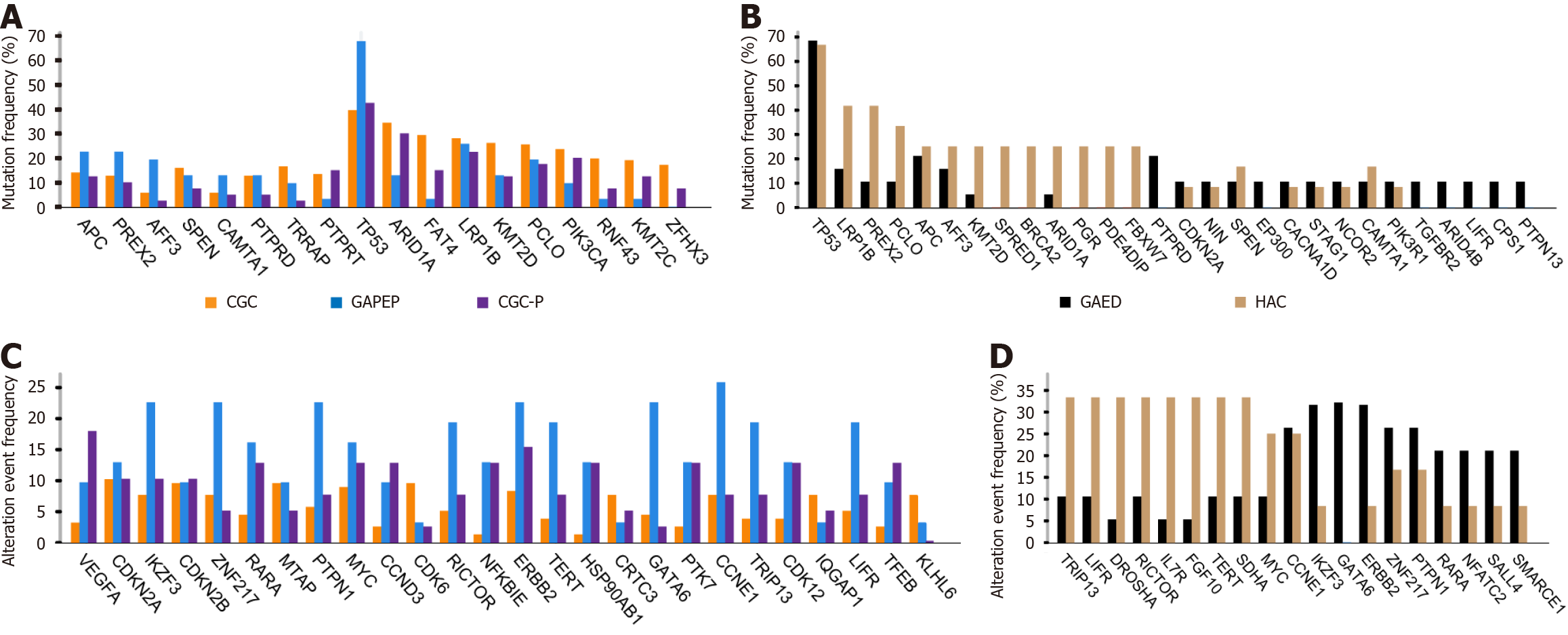
Figure 4 Distinct genomic landscapes across gastric cancer subtypes.
A: Somatic mutation analysis identifying the top 10 most frequently mutated genes in gastric adenocarcinoma with primitive enterocyte phenotype (blue), conventional gastric cancer (CGC) (orange) cohorts and CGC expressing primitive phenotypic markers (purple), with mutation frequencies expressed as percentages; B: Somatic mutation analysis revealing the top 10 most frequently mutated genes in hepatoid adenocarcinoma (brown) and gastric adenocarcinoma with enteroblastic differentiation (black) cohorts, with both groups exhibiting notably high TP53 mutation frequencies; C: Copy number alteration analysis highlighting the top 10 genes with the highest alteration frequencies in gastric adenocarcinoma with primitive enterocyte phenotype (blue), CGC expressing primitive phenotypic markers (purple), and CGC (orange) cohorts; D: Copy number alteration analysis showcasing the top 10 genes with the highest alteration frequencies in hepatoid adenocarcinoma (brown) and gastric adenocarcinoma with enteroblastic differentiation (black) cohorts. GAPEP: Gastric adenocarcinoma with primitive enterocyte phenotype; CGC: Conventional gastric cancer; CGC-P: Conventional gastric cancer expressing primitive phenotypic markers; HAC: Hepatoid adenocarcinoma; GAED: Gastric adenocarcinoma with enteroblastic differentiation.
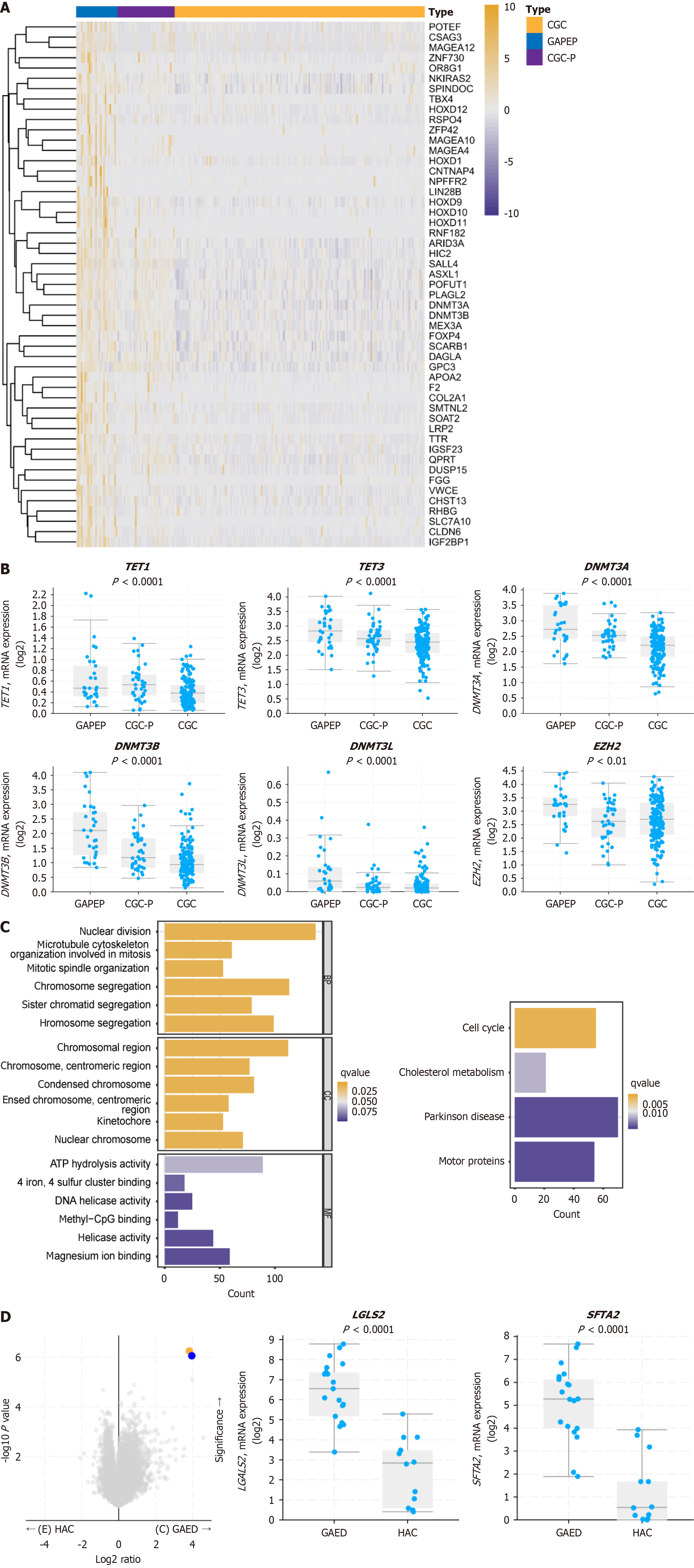
Figure 5 Multi-omics profiling reveals methylation-driven molecular features of gastric adenocarcinoma with primitive enterocyte phe
- Citation: Li HQ, Zheng LQ, Huang WT, Yu XB, Zhang X, Lin L, Lv SS, Yan XY, Chen XY. Clinicopathological significance of histological diversity in gastric adenocarcinoma with primitive enterocyte phenotype: A methylation-driven aggressive entity. World J Gastroenterol 2025; 31(28): 108990
- URL: https://www.wjgnet.com/1007-9327/full/v31/i28/108990.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i28.108990