©The Author(s) 2023.
World J Gastroenterol. Apr 14, 2023; 29(14): 2064-2077
Published online Apr 14, 2023. doi: 10.3748/wjg.v29.i14.2064
Published online Apr 14, 2023. doi: 10.3748/wjg.v29.i14.2064
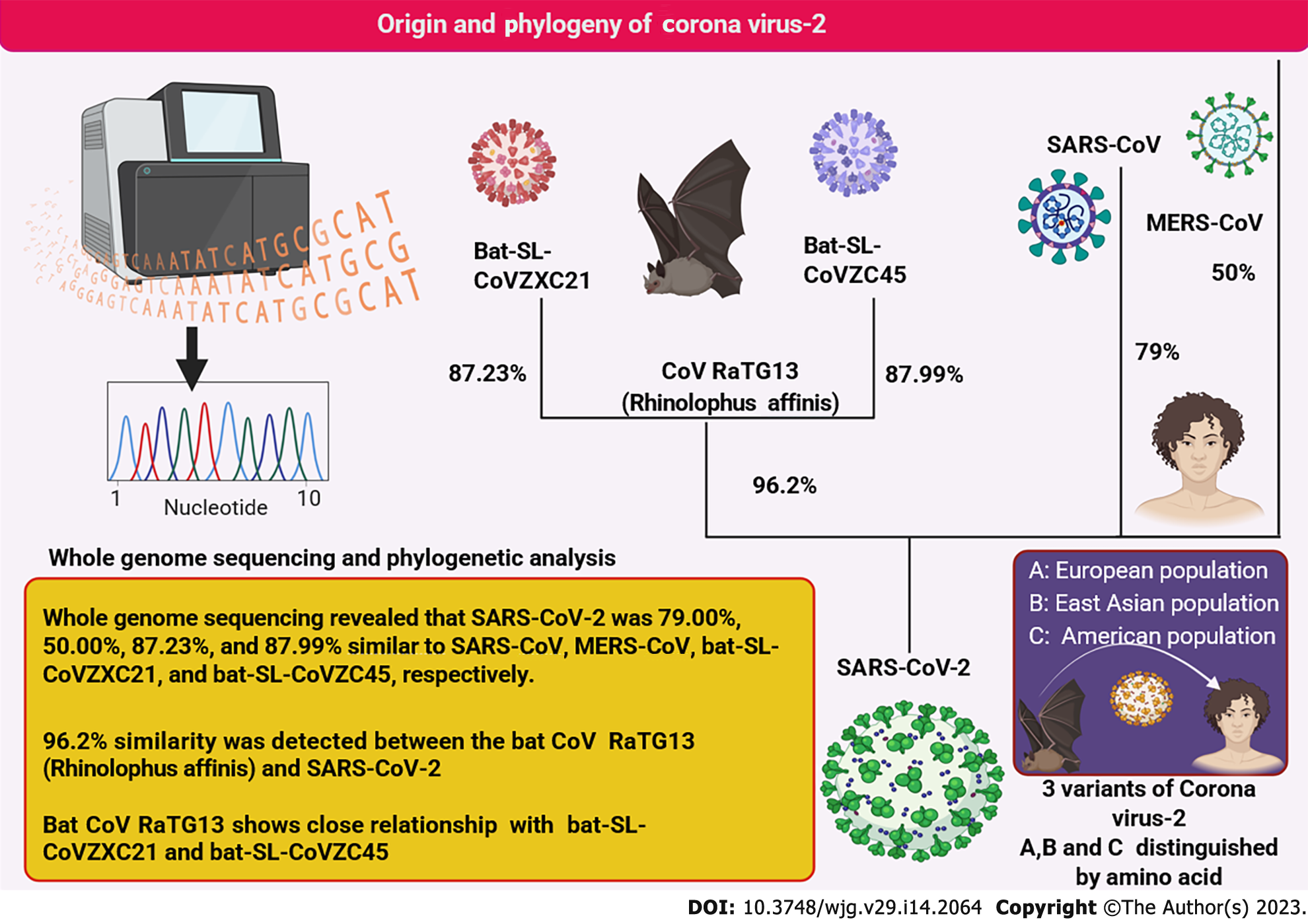
Figure 1 The origin and ancestral relationship of severe acute respiratory syndrome coronavirus 2 with other related viruses.
Whole genome sequencing revealed that severe acute respiratory syndrome coronavirus 2 virus was most closely related to the CoV RaTG13 virus. The variant strains of coronavirus and their biogeographical distribution are illustrated. MERS-CoV: Middle East respiratory syndrome coronavirus; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; R. affinis RaTG13: SARS-like coronaviruses: Bat-SLCoVZX.
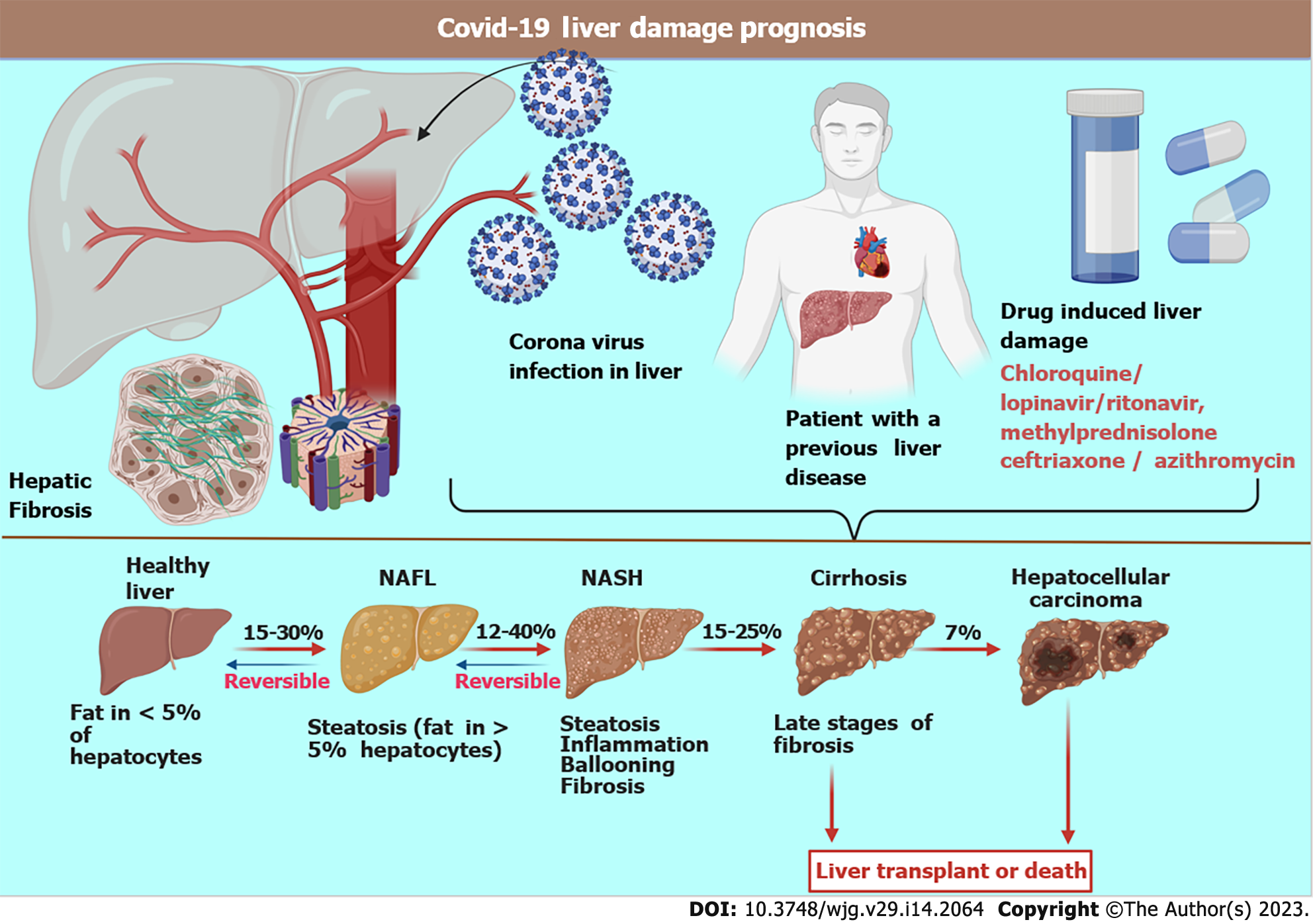
Figure 2 The schematic representation of the effects of severe acute respiratory syndrome coronavirus 2 on the liver.
A patient with a pre-existing liver disorder exhibits higher morbidity than patients with normal liver. Drugs may cause liver damage in some patients, making treatments ineffective. The stages of disease progression are implicated as steatosis (deposition of fat), causing a nonalcoholic fatty liver. The liver deteriorates, indicated by inflammation, ballooning of hepatocytes, and fibrosis. Severe liver cirrhosis is the next stage, which is irreversible. The final stage is hepatic cancer. COVID-19: Coronavirus disease 2019; NAFL: Nonalcoholic fatty liver; NASH: Nonalcoholic steatohepatitis.
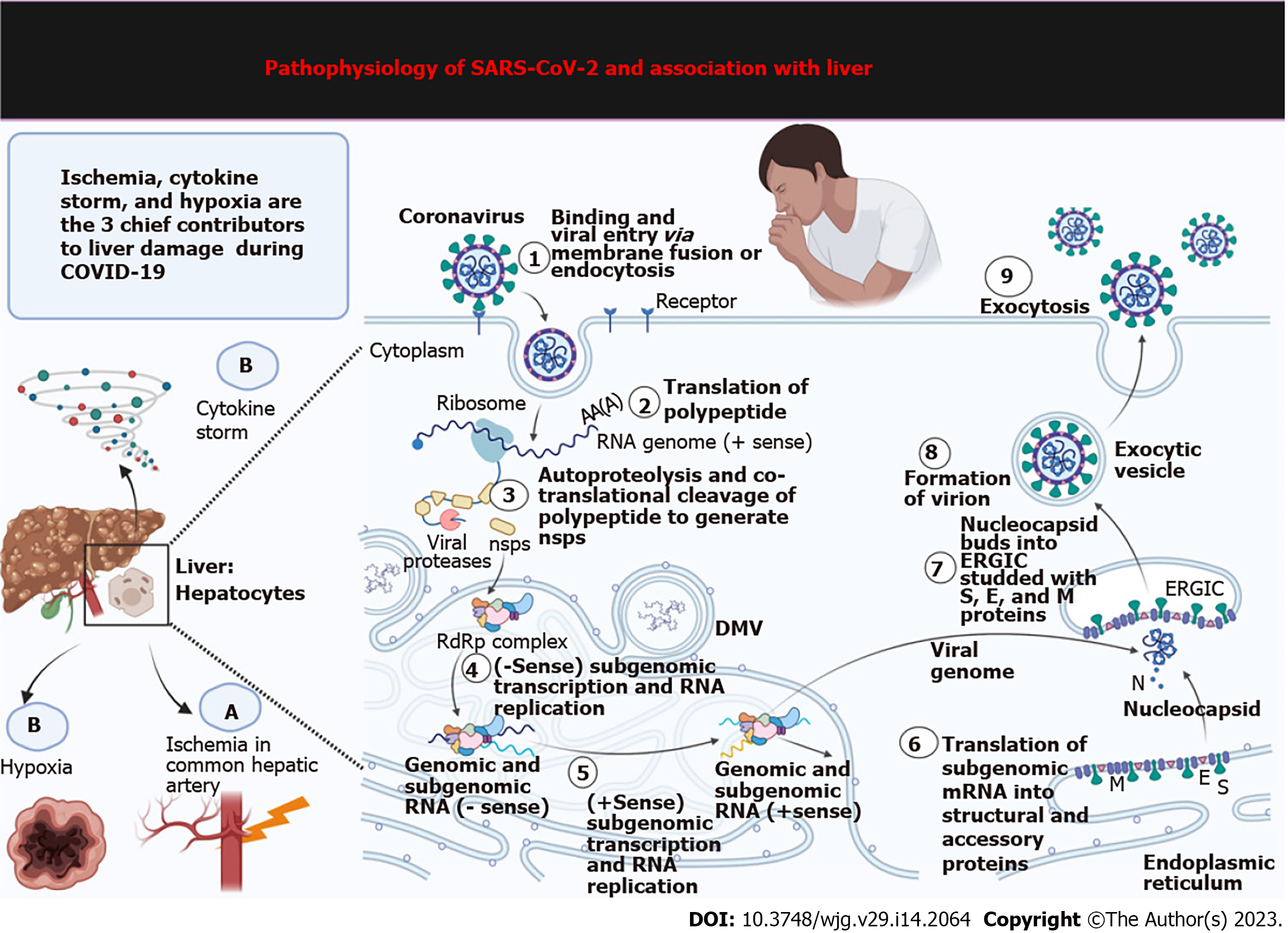
Figure 3 Molecular mechanisms of liver damage during coronavirus disease 2019 infection.
Hypoxia, ischemia, and cytokine storm significantly contribute to liver damage in coronavirus disease 2019-related comorbidities. This figure depicts how severe acute respiratory syndrome coronavirus 2 exponentially proliferates after entering hepatocytes. The steps occur: (1) Binding and viral entry via membrane fusion or endocytosis; (2) Translation of polypeptide; (3) Autoproteolysis and cotranslational polypeptide cleavage to generate non-structural proteins (nsps); (4) Sense subgenomic transcription and RNA replication; (5) + Sense subgenomic transcription and RNA replication; (6) Translation of subgenomic mRNA into structural and accessory proteins; (7) Nucleocapsid buds into ER-Golgi intermediate compartment studded with spike, envelope, and membrane proteins; (8) Formation of virion; and (9) Exocytosis. SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; COVID-19: Coronavirus disease 2019; ERGIC: ER-Golgi intermediate compartment; DMV: Double-membrane vesicle.
- Citation: Khullar N, Bhatti JS, Singh S, Thukral B, Reddy PH, Bhatti GK. Insight into the liver dysfunction in COVID-19 patients: Molecular mechanisms and possible therapeutic strategies. World J Gastroenterol 2023; 29(14): 2064-2077
- URL: https://www.wjgnet.com/1007-9327/full/v29/i14/2064.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i14.2064