©The Author(s) 2022.
World J Gastroenterol. Aug 28, 2022; 28(32): 4681-4697
Published online Aug 28, 2022. doi: 10.3748/wjg.v28.i32.4681
Published online Aug 28, 2022. doi: 10.3748/wjg.v28.i32.4681
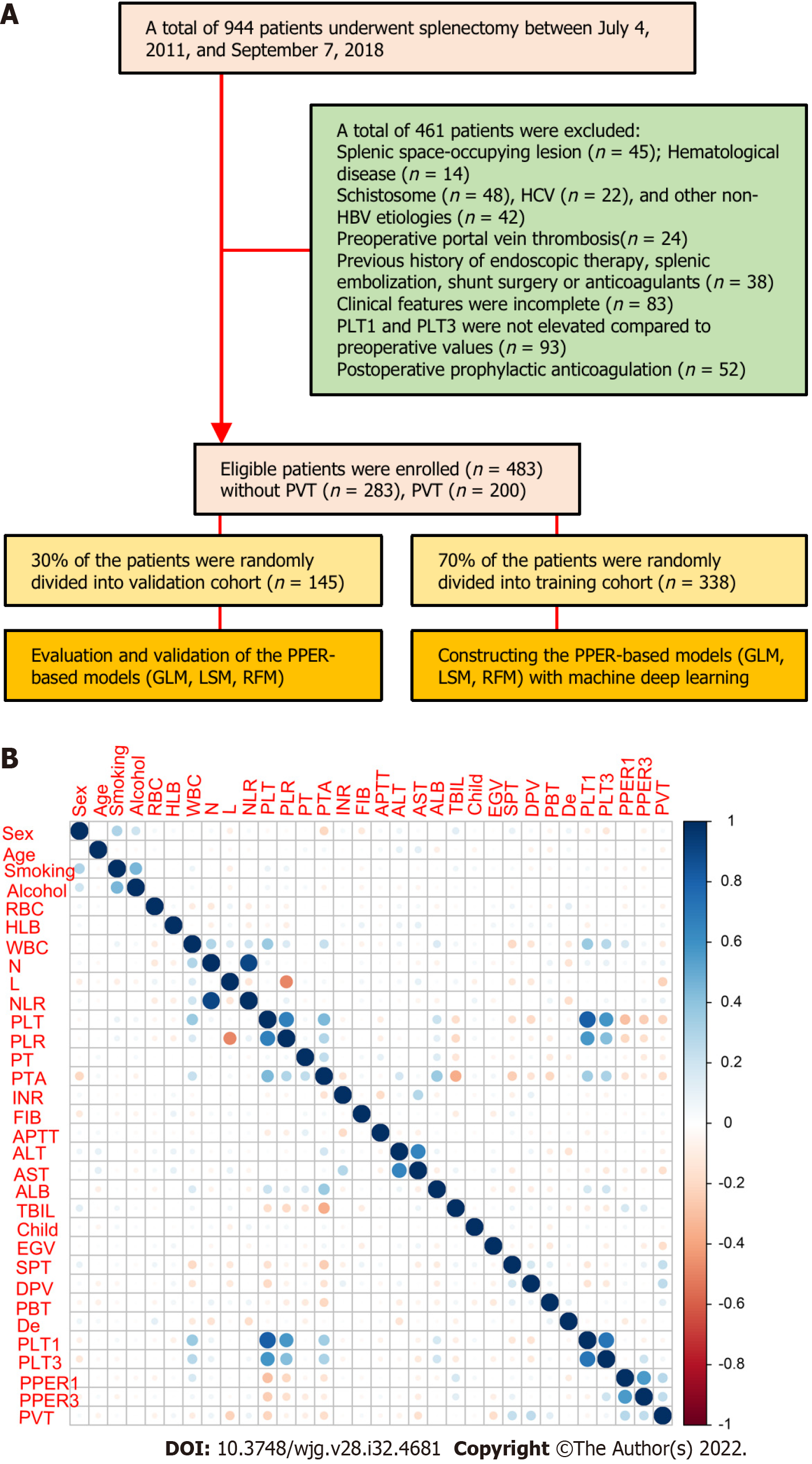
Figure 1 Flow chart and correlation chart.
A: Flow diagram of patient selection and study design; B: Correlation matrix between candidate variables. The size and color of the circle in the matrix reflect the correlation between the corresponding variables. The darker the blue, the stronger the positive correlation between variables, and the darker the red, the stronger the negative correlation between variables. HCV: Hepatitis C virus; HBV: Hepatitis B virus; PLT1 and PLT3: Platelet counts on the first and third days after operation; PVT: Portal vein thrombosis; GLM: Generalized linear model; LSM: Least absolute shrinkage and selection operator model; RFM: Random forest model; RBC: Red blood cells; HLB: Hemoglobin; WBC: White blood cells; N: Neutrophil count; L: Lymphocyte count; NLR: Neutrophil to lymphocyte ratio; PLT: Platelet count; PLR: Platelet to lymphocyte ratio; PT: Prothrombin time; PTA: Prothrombin activity; INR: International normalized ratio; FIB: Fibrinogen; APTT: Activated partial thromboplastin time; ALT: Alanine aminotransaminase; AST: Aspartate aminotransaminase; ALB: Serum albumin; TBIL: Total serum bilirubin; Child: Child-Pugh grade; EGV: Esophageal and gastric varices; SPT: Spleen thickness; DPV: Diameter of the portal vein; PBT: Preoperative blood transfusion; De: Devascularization; PPER1: Platelet elevation rate at postoperative day 1; PPER3: Platelet elevation rate at postoperative day 3.
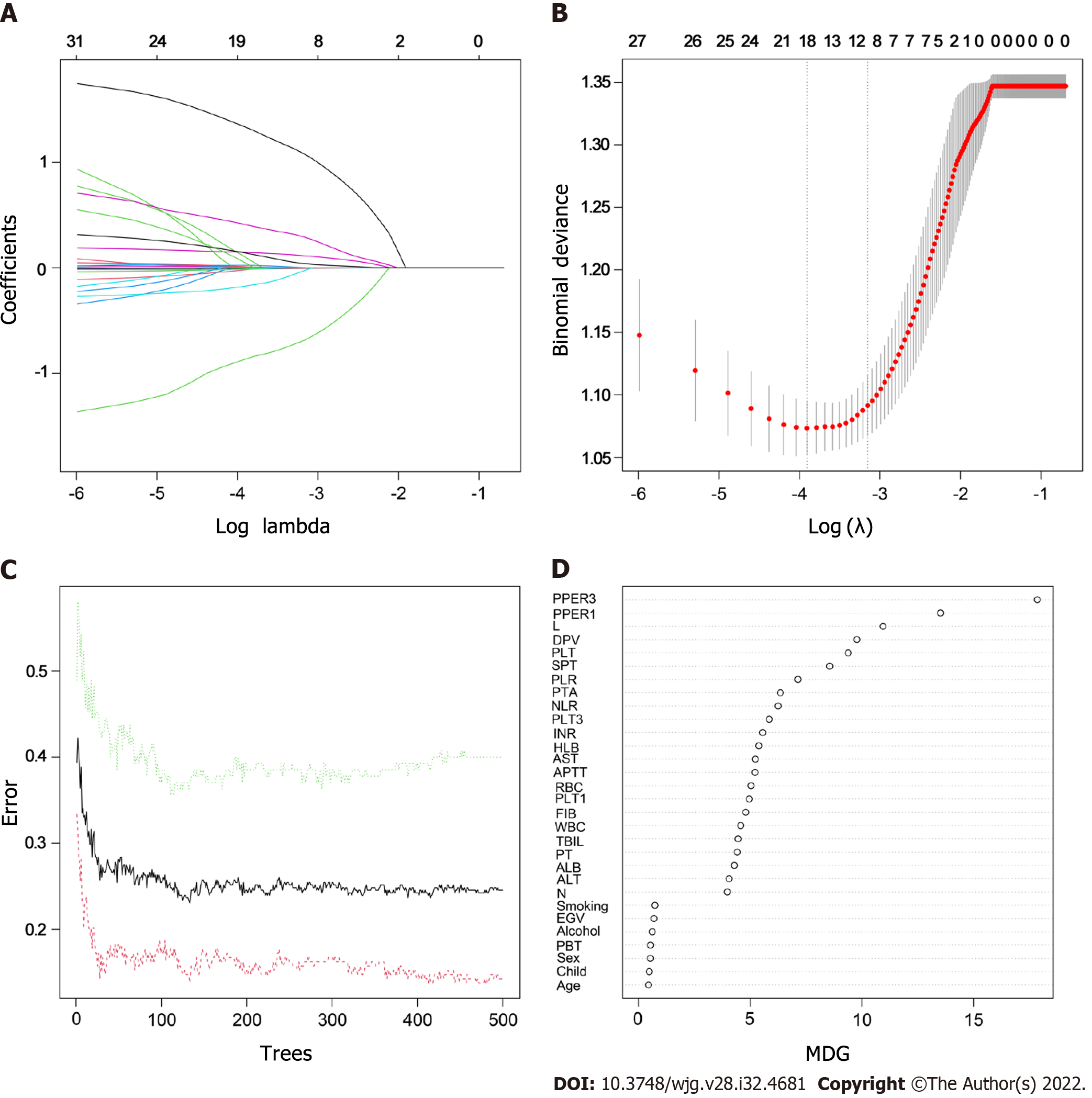
Figure 2 Features selection.
A: Least absolute shrinkage and selection operator variable trace profiles of the 31 features. The 3-fold cross-validation was employed; B: Mean square error (MSE) plots of models under different lambda. The lambda corresponding to one MSE away from the minimum MSE was the optimal lambda value of 0.043, and the target variables shrunk to 10; C: Relationships between the error and number of trees. There are three lines, green representing error in the positive event group, red representing error in the negative event group, and black representing error in the total sample group; D: Importance scores of the candidate features. RBC: Red blood cells; HLB: Hemoglobin; WBC: White blood cells; N: Neutrophil count; L: Lymphocyte count; NLR: Neutrophil to lymphocyte ratio; PLT: Platelet count; PLR: Platelet to lymphocyte ratio; PT: Prothrombin time; PTA: Prothrombin activity; INR: International normalized ratio; FIB: Fibrinogen; APTT: Activated partial thromboplastin time; ALT: Alanine aminotransaminase; AST: Aspartate aminotransaminase; ALB: Serum albumin; TBIL: Total serum bilirubin; Child: Child-Pugh grade; EGV: Esophageal and gastric varices; SPT: Spleen thickness; DPV: Diameter of the portal vein; PBT: Preoperative blood transfusion; PLT1: Platelet count at postoperative day 1; PLT3: Platelet count at postoperative day 3; PPER1: Platelet elevation rate at postoperative day 1; PPER3: Platelet elevation rate at postoperative day 3; MDG: Mean decreased Gini.
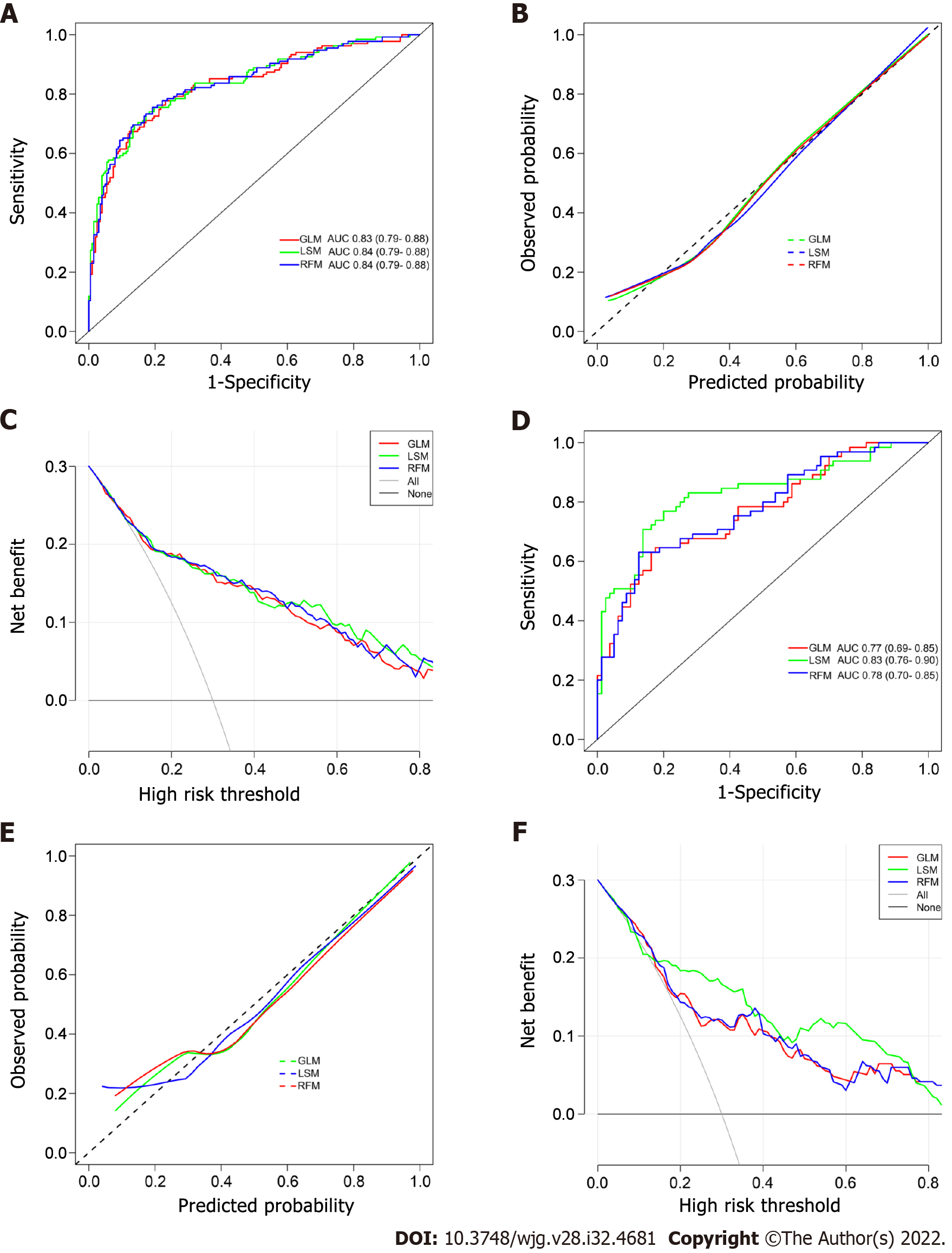
Figure 3 Evaluation and validation of the postoperative platelet elevation rate-based models in the training cohort and validation cohort.
A and D: The receiver operating characteristic curves of the postoperative platelet elevation rate (PPER)-based models; B and E: The calibration curves of the PPER-based models; C and F: The decision curve analysis of the PPER-based models. GLM: Generalized linear model; LSM: Least absolute shrinkage and selection operator model; RFM: Random forest model; AUC: Area under the receiver operating characteristic curve.
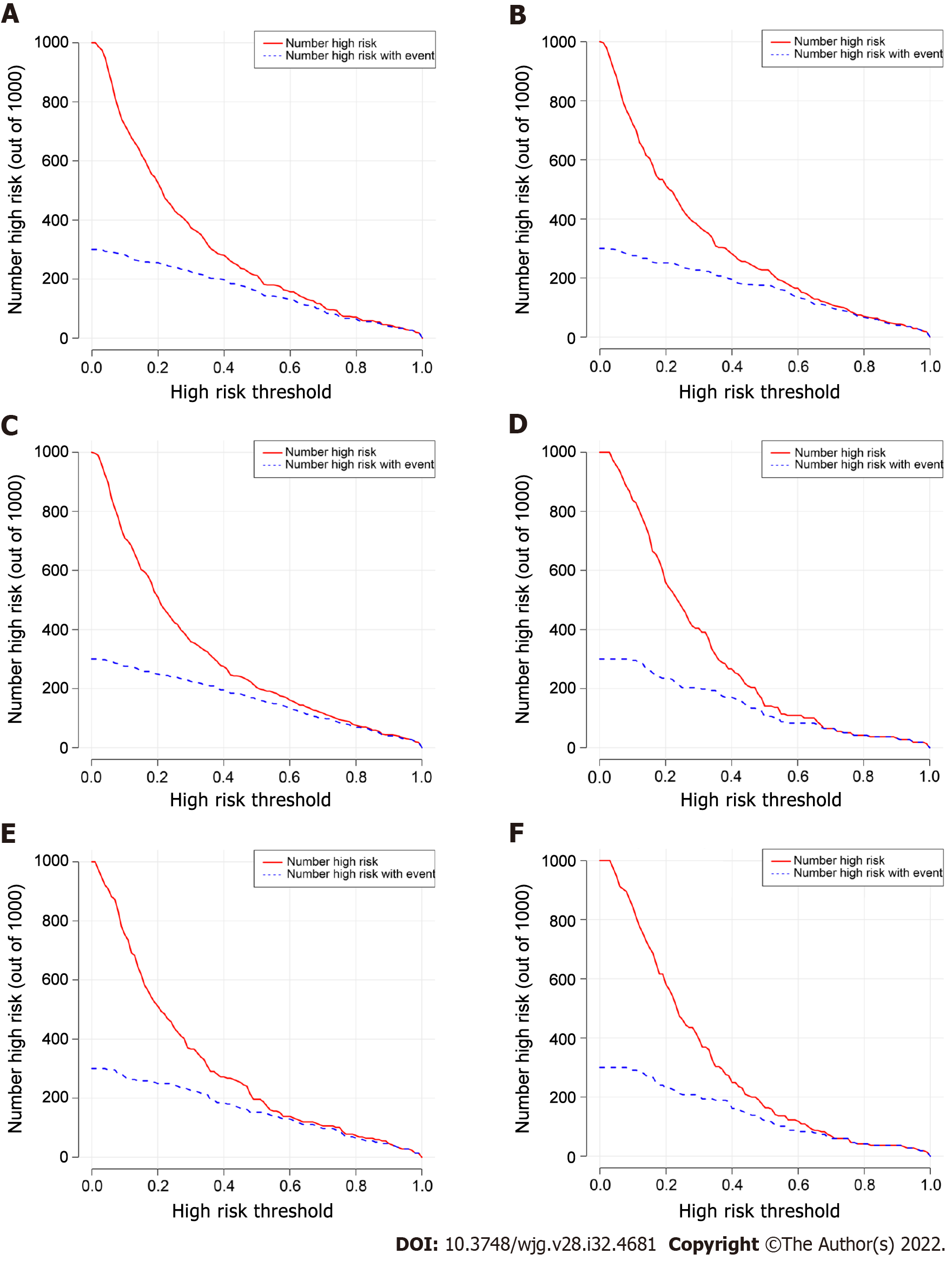
Figure 4 Clinical impact curves of the postoperative platelet elevation rate-based models in the training cohort and validation cohort.
A and D: Clinical impact curves for the generalized linear model; B and E: Clinical impact curves for the least absolute shrinkage and selection operator model; C and F: Clinical impact curves for the random forest model.
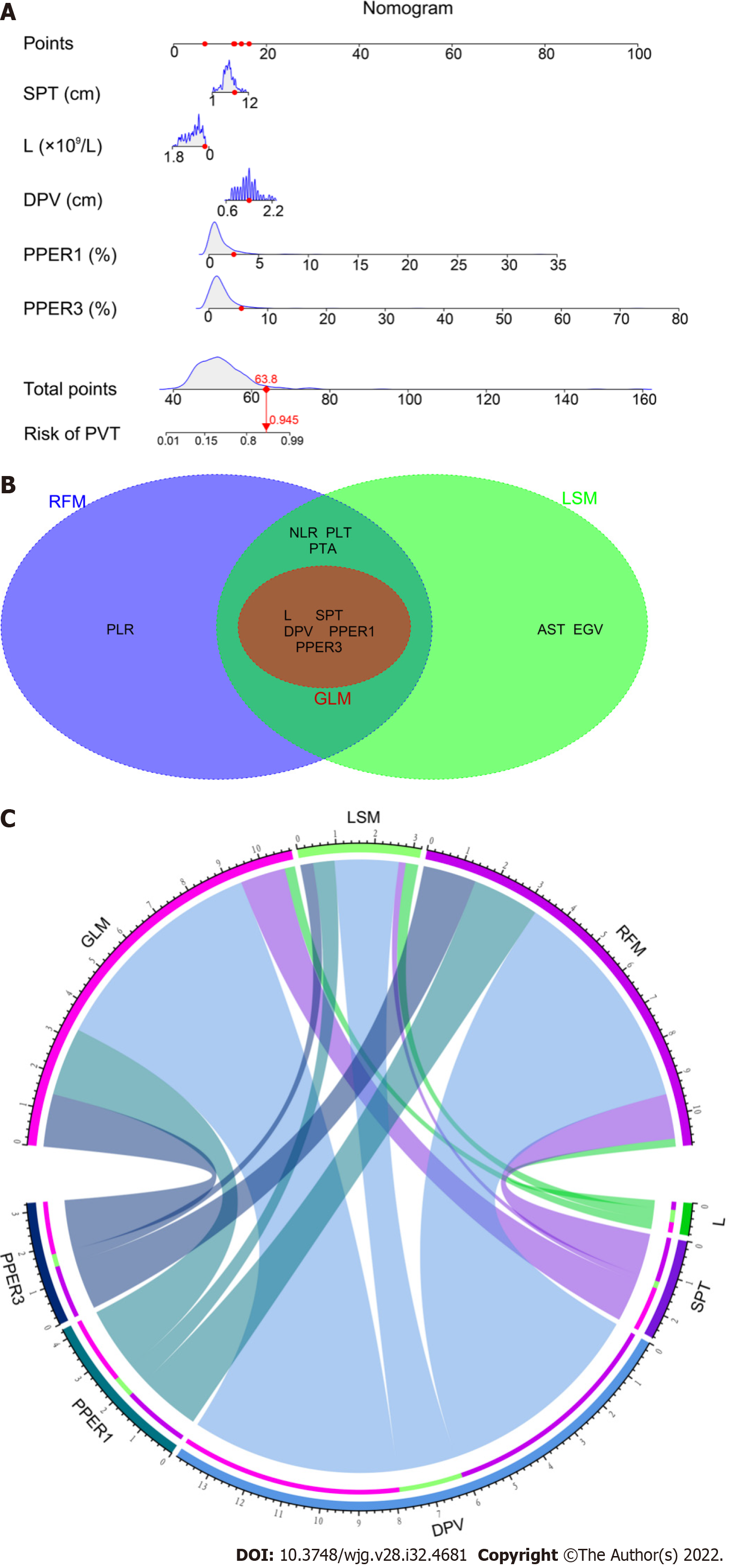
Figure 5 Nomogram for the generalized linear model and weighting of variables.
A: Nomogram for the generalized linear model (GLM); B: Intersection variables among the GLM, least absolute shrinkage and selection operator model (LSM), and random forest model (RFM); C: Weights of the intersection variables in the GLM, LSM, and RFM, respectively. SPT: Spleen thickness; L: Lymphocyte count; DPV: Diameter of portal vein; PPER1 and PPER3: The first and third days for postoperative platelet elevation rate; PVT: Portal vein thrombosis; PLR: Platelet to lymphocyte ratio; NLR: Neutrophil to lymphocyte ratio; PLT: Platelet count; PTA: Prothrombin activity; EGV: Esophageal and gastric varices; AST: Aspartate aminotransaminase.
- Citation: Li J, Wu QQ, Zhu RH, Lv X, Wang WQ, Wang JL, Liang BY, Huang ZY, Zhang EL. Machine learning predicts portal vein thrombosis after splenectomy in patients with portal hypertension: Comparative analysis of three practical models. World J Gastroenterol 2022; 28(32): 4681-4697
- URL: https://www.wjgnet.com/1007-9327/full/v28/i32/4681.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i32.4681