Copyright
©The Author(s) 2020.
World J Gastroenterol. Jun 28, 2020; 26(24): 3401-3412
Published online Jun 28, 2020. doi: 10.3748/wjg.v26.i24.3401
Published online Jun 28, 2020. doi: 10.3748/wjg.v26.i24.3401
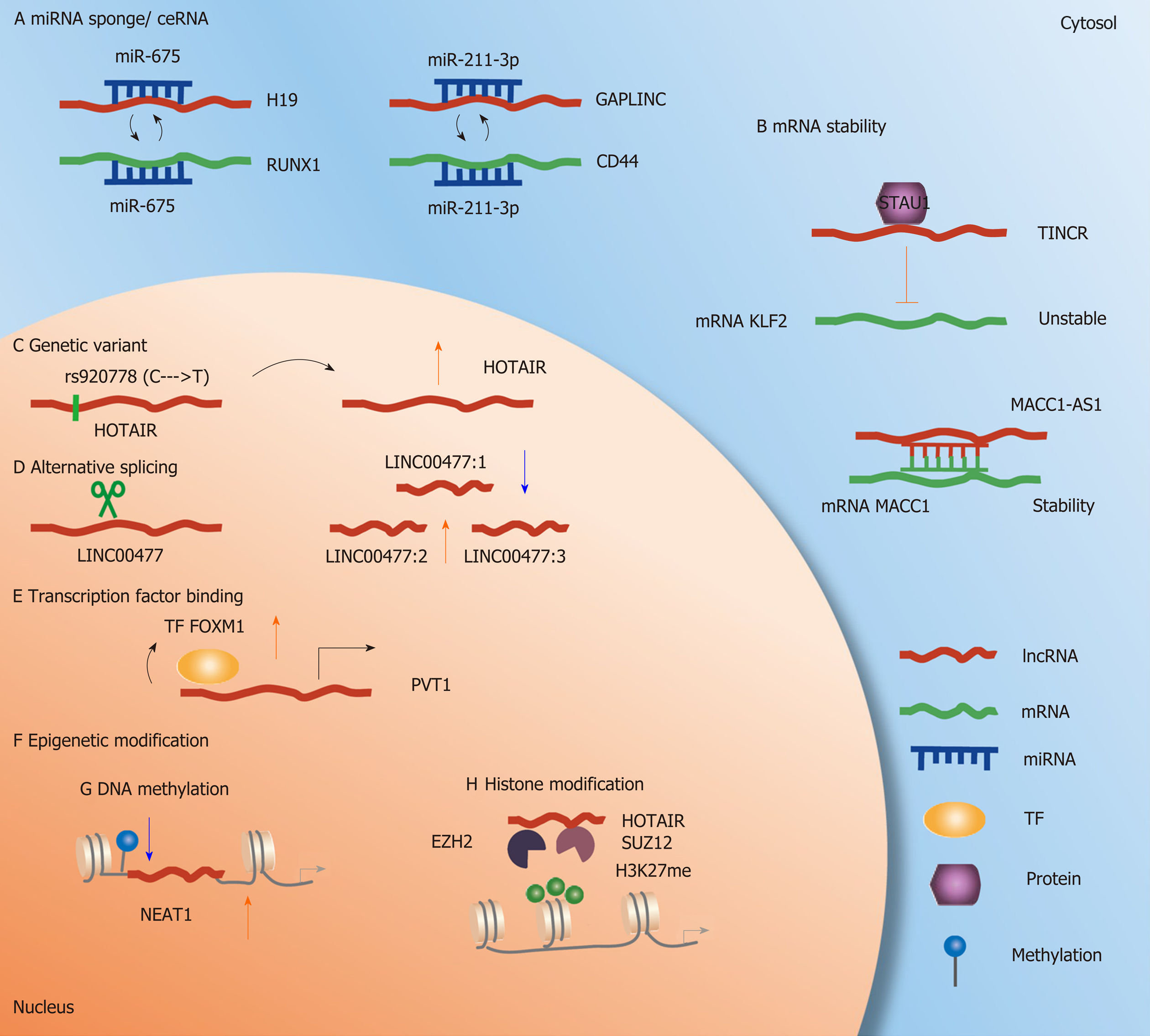
Figure 1 Molecular mechanism of long noncoding RNAs in gastric cancer.
A: MicroRNA (miRNA) sponge. H19 and GAPLINC, acting as molecular sponges, bind to miR-675 and miR-211-3p, thus increasing the expression of their target genes RUNX1 and Cd44; B: mRNA stability. Target transcripts are stabilized or decayed when long noncoding RNA (lncRNA) binds to mRNA; C: Genetic variants. Genetic variants located at lncRNAs influence the expression of lncRNA; D: Alternative splicing. LINC00477 generates three transcripts based on alternative splicing; E: Transcription factor binding. Transcription factor directly interacts with the lncRNA PVT1 and regulates its expression; F: Epigenetic modification; G: DNA methylation. DNA methylation sites located at or near lncRNA regulate lncRNA expression; H: Histone modification. H3K27me results in suppression or gain of transcriptional activity.
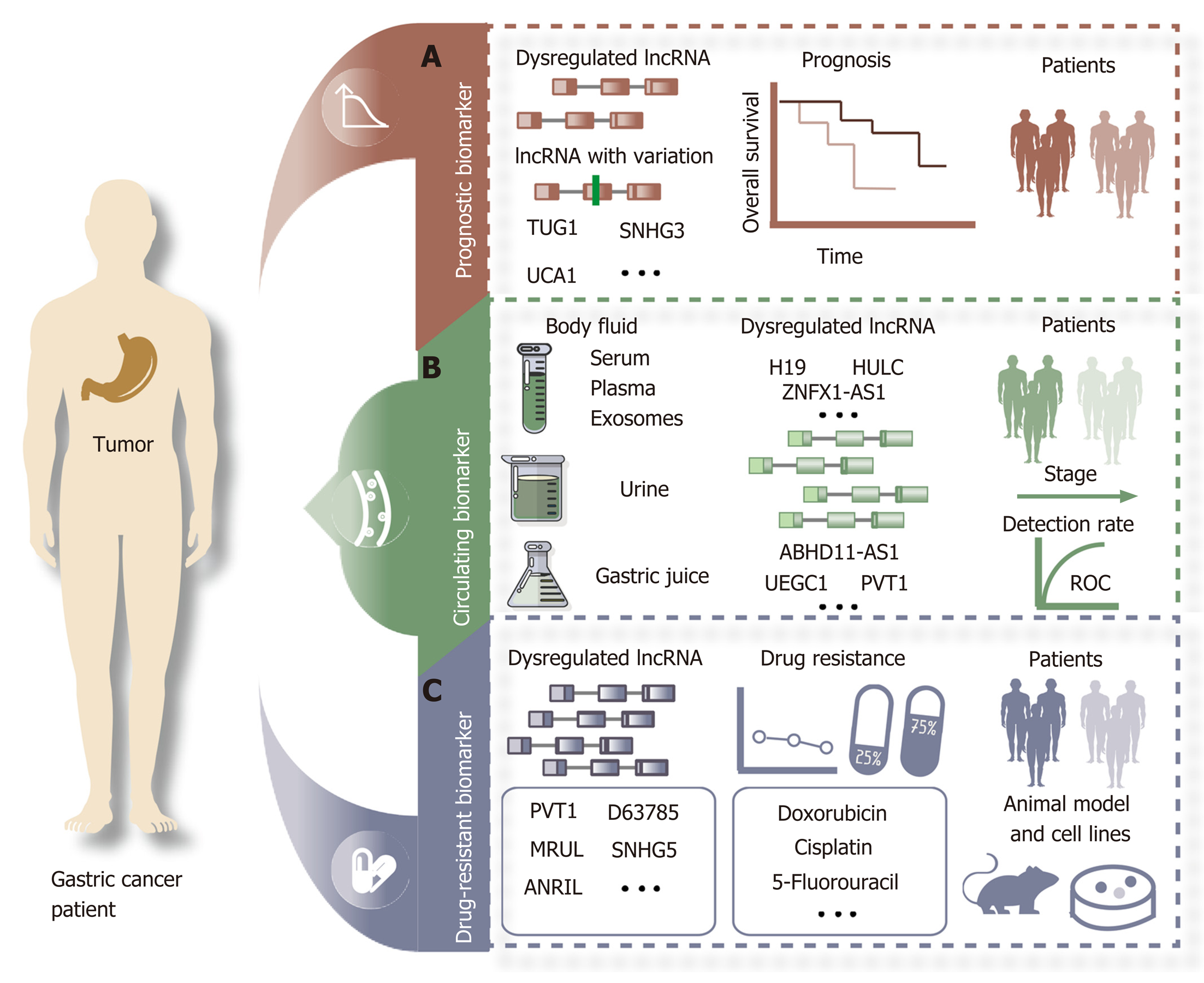
Figure 2 Potential clinical application of long noncoding RNAs as biomarkers in gastric cancer.
A: Long noncoding RNAs (lncRNAs) with dysregulated expression or variants can act as prognostic biomarkers for gastric cancer (GC) patients; B: Dysregulated lncRNAs identified from body fluids can be used to detect GC; C: Selected dysregulated lncRNAs are associated with drug resistance in GC patients, animal models, and cell lines. lncRNA: Long noncoding RNA.
- Citation: Gao Y, Wang JW, Ren JY, Guo M, Guo CW, Ning SW, Yu S. Long noncoding RNAs in gastric cancer: From molecular dissection to clinical application. World J Gastroenterol 2020; 26(24): 3401-3412
- URL: https://www.wjgnet.com/1007-9327/full/v26/i24/3401.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i24.3401