©The Author(s) 2016.
World J Gastroenterol. Jan 21, 2016; 22(3): 1190-1201
Published online Jan 21, 2016. doi: 10.3748/wjg.v22.i3.1190
Published online Jan 21, 2016. doi: 10.3748/wjg.v22.i3.1190
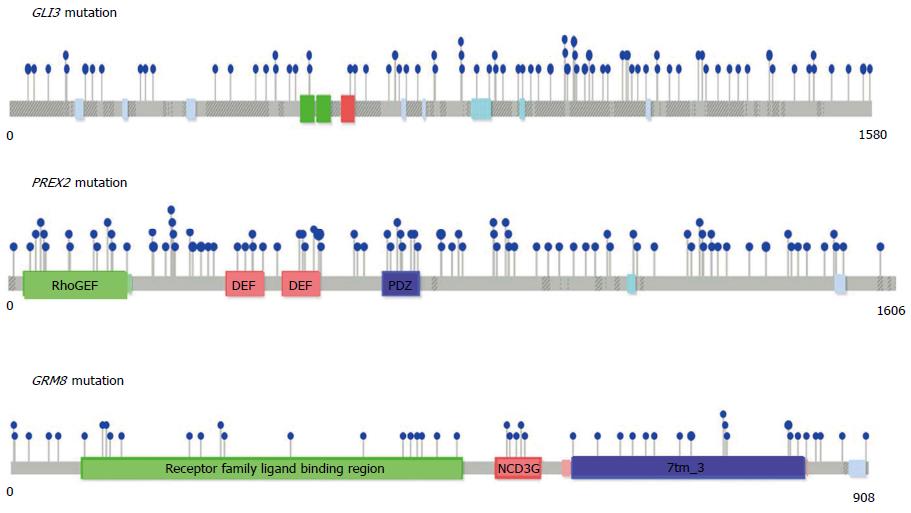
Figure 1 Mutational positions in three significantly mutated genes, GLI3, PREX2, and GRM8.
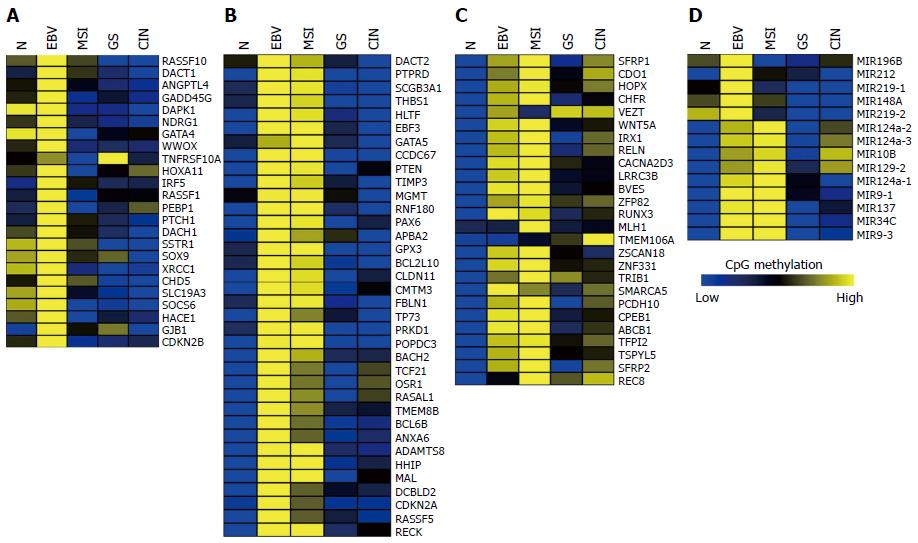
Figure 2 Methylation profiles of the previously reported 86 genes and 14 miRNAs analyzed using the Cancer Genome Atlas methylation data.
A: Genes hypermethylated in Epstein-Barr virus (EBV)-positive gastric cancers (GCs); B: Genes hypermethylated in both EBV-positive and MSI-high GCs; C: Genes hypermethylated in more than two GC subtypes; D: Methylation profiles of miRNAs in four GC subtypes.
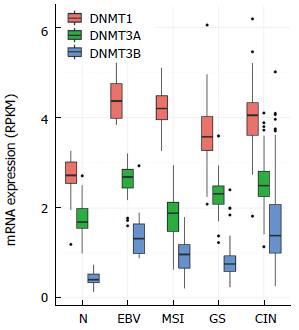
Figure 3 Expression levels of DNMT1, DNMT3A, and DNMT3B analyzed using the Cancer Genome Atlas RNA sequencing data.
EBV: Epstein-Barr virus.
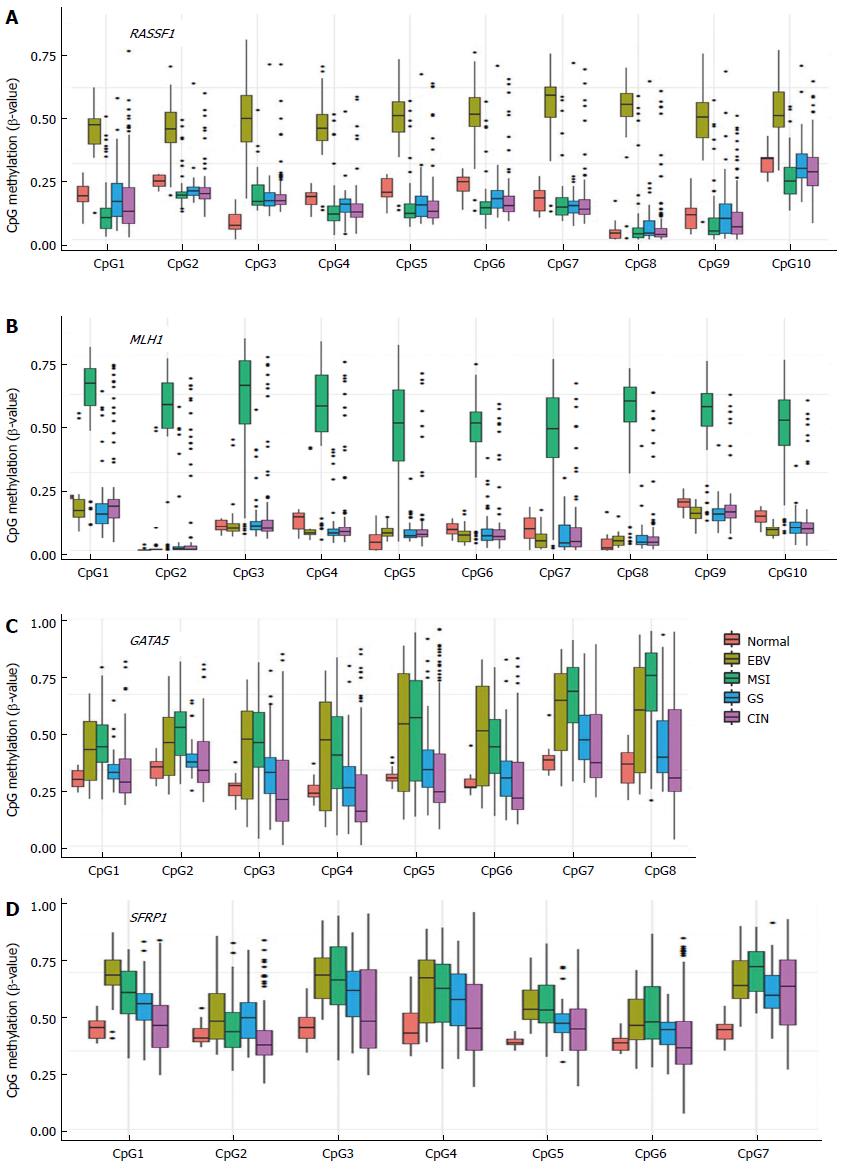
Figure 4 CpG methylation level in promoter regions of RASSF1 (A), MLH1 (B), GATA5 (C), and SFRP1 (D) analyzed using the Cancer Genome Atlas methylation data.
- Citation: Lim B, Kim JH, Kim M, Kim SY. Genomic and epigenomic heterogeneity in molecular subtypes of gastric cancer. World J Gastroenterol 2016; 22(3): 1190-1201
- URL: https://www.wjgnet.com/1007-9327/full/v22/i3/1190.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i3.1190