©The Author(s) 2015.
World J Gastroenterol. Apr 7, 2015; 21(13): 3960-3969
Published online Apr 7, 2015. doi: 10.3748/wjg.v21.i13.3960
Published online Apr 7, 2015. doi: 10.3748/wjg.v21.i13.3960
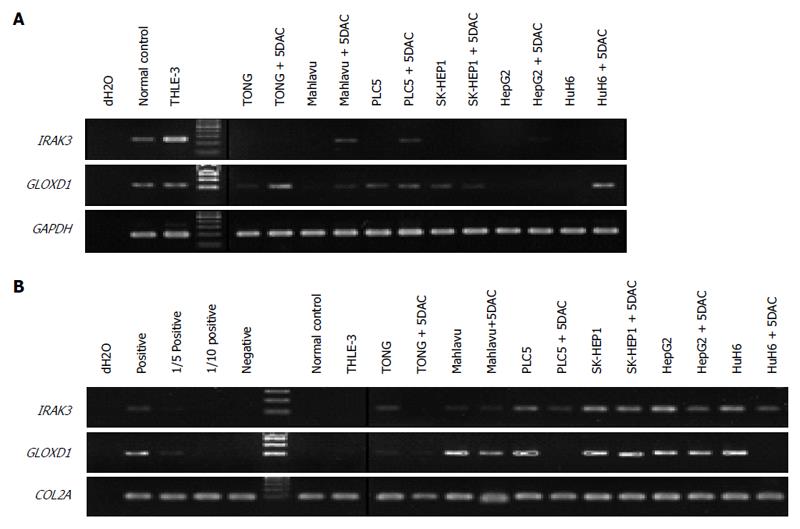
Figure 1 Gene expression and methylation analyses of IRAK3 and GLOXD1.
A: Gene expression levels of IRAK3, GLOXD1, and GAPDH (an internal reference gene) were analyzed by RT-PCR in normal controls, THLE-3 cells, 6 HCC cell lines, and HCC cell lines treated with 5DAC; B: Methylation status of IRAK3, GLOXD1, and COL2A (an internal reference gene) was analyzed by MS-PCR with methylated primers in normal controls, THLE-3 cells, 6 HCC cell lines, and HCC cell lines treated with 5DAC. Positive and negative are peripheral blood lymphocyte (PBL) DNA in vitro treated with or without CpG methyltransferase (M.SssI). 1/5 positive and 1/10 positive indicate 1:5 and 1:10 dilution of the positive control.
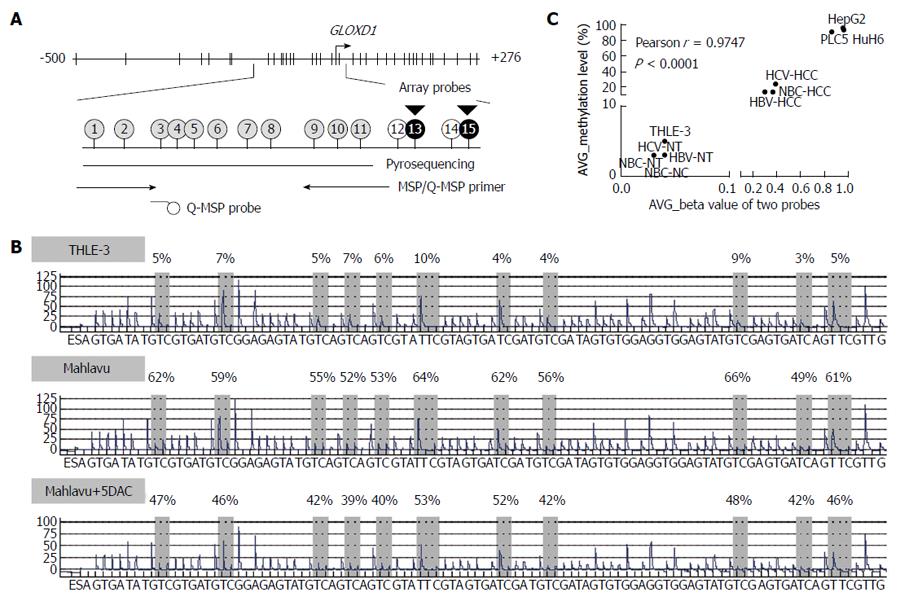
Figure 2 Map of the GLOXD1 promoter region and representative methylation pattern determined by pyrosequencing.
A: The 15 CpG sites within the GLOXD1 promoter (-500/+276) were addressed using different techniques. Two black circles indicate the 2 CpG sites recognized by probes of the methylation array chip, respectively. Eleven gray circles indicate the 11 CpG sites addressed by pyrosequencing and the 6 CpG sites that MSP/Q-MSP primer set covered (5 CpG sites for allele-specific, one CpG site for probe); B: Methylation level of 11 CpG sites addressed by pyrosequencing in THLE-3 cells, Mahlavu cells, and Mahlavu cells treated with 5DAC; C: Pearson correlation was analyzed between the average β value of two array probes and average methylation levels of the 11 CpG sites assessed by pyrosequencing in samples for methylation array, including 4 cell lines and 7 types of liver tissues.
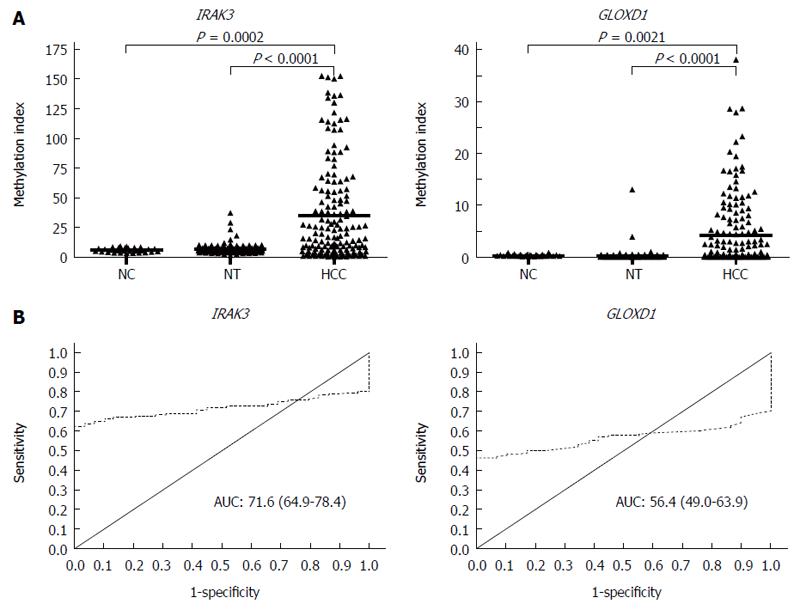
Figure 3 Methylation levels and receiver operating characteristic curve analysis of IRAK3 and GLOXD1 in liver tissues.
A: Gene methylation was determined in 29 normal controls (NC) and 160 paired hepatocellular carcinoma (HCC) tissues and their adjacent notumor tissues (NT) by quantitative methylation-specific polymerase chain reaction. The results are represented as the difference in the methylation index. The black lines indicate the mean of the methylation index. (NC vs HCC, unpaired t-test; NT vs HCC, paired t-test); B: The area under the receiver operating characteristic curve (AUC) for each gene was calculated to discriminate 29 normal individuals and 160 HCC cases.
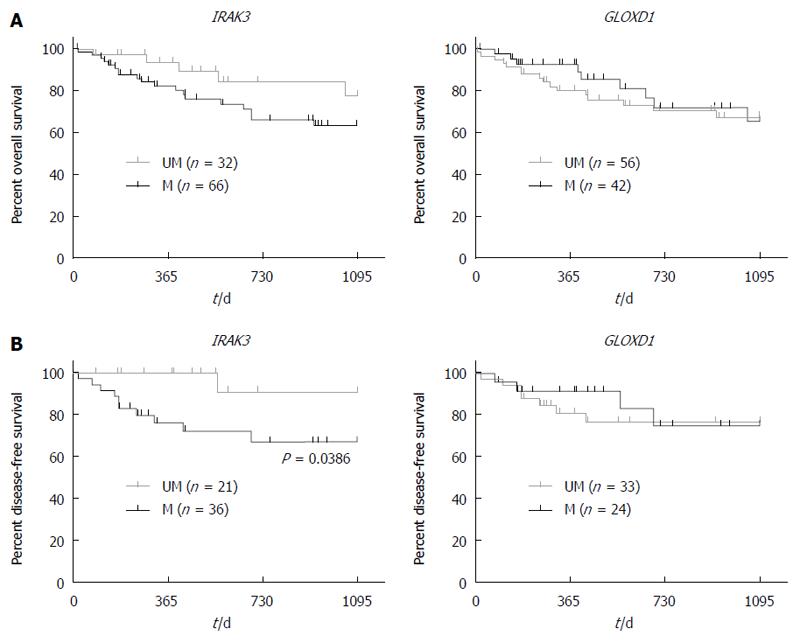
Figure 4 Correlation analyses between gene methylation and the survival of hepatocellular carcinoma patients.
A: Survival was analyzed using Kaplan-Meier curves. The plots were made according to the patients with IRAK3 and GLOXD1 methylation and 3-year overall survival in 98 hepatocellular carcinoma (HCC) patients, respectively; B: Kaplan-Meier survival curves were made according to the cases with GLOXD1 and IRAK3 methylation and 3-year disease-free survival in 57 HCC patients (P = 0.0386, log-rank test, UM, unmethylated cases vs M, methylated cases).
-
Citation: Kuo CC, Shih YL, Su HY, Yan MD, Hsieh CB, Liu CY, Huang WT, Yu MH, Lin YW. Methylation of
IRAK3 is a novel prognostic marker in hepatocellular carcinoma. World J Gastroenterol 2015; 21(13): 3960-3969 - URL: https://www.wjgnet.com/1007-9327/full/v21/i13/3960.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i13.3960