©2014 Baishideng Publishing Group Co.
World J Gastroenterol. Feb 28, 2014; 20(8): 2042-2050
Published online Feb 28, 2014. doi: 10.3748/wjg.v20.i8.2042
Published online Feb 28, 2014. doi: 10.3748/wjg.v20.i8.2042
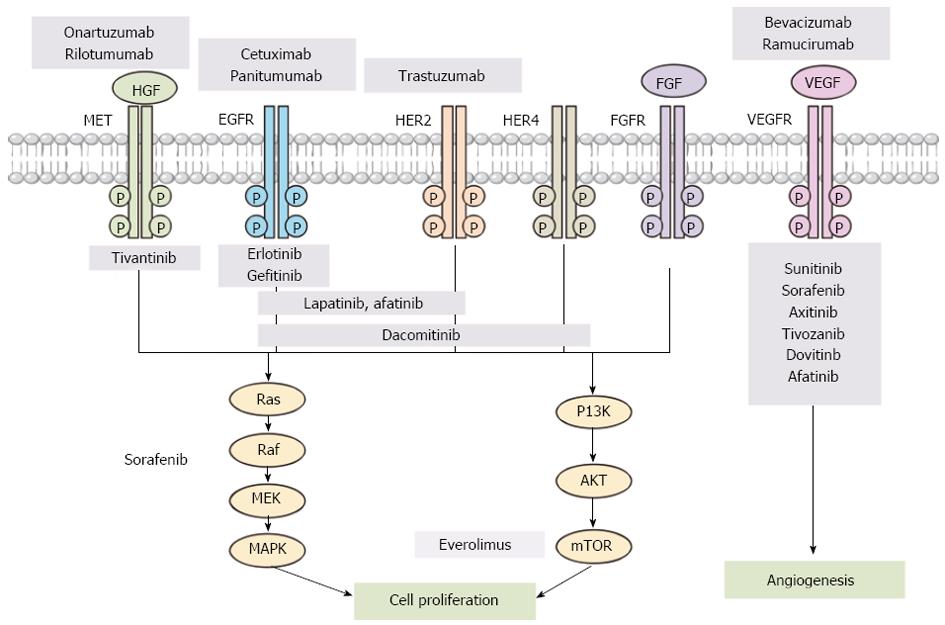
Figure 1 Molecular targeting agents for different signaling pathways in gastric cancer.
PI3K: Phosphatidylinositol 3-kinase; HER: Human epidermal growth factor receptor; FGFR: Fibroblast growth factor (FGF) receptor; mTOR: Mammalian target of rapamycin; HGF: Hepatocyte growth factor; VEGFR: Vascular endothelial growth factor (VEGF) receptor; EGF: Epidermal growth factor; MAPK: Mitogen-activated protein kinase.
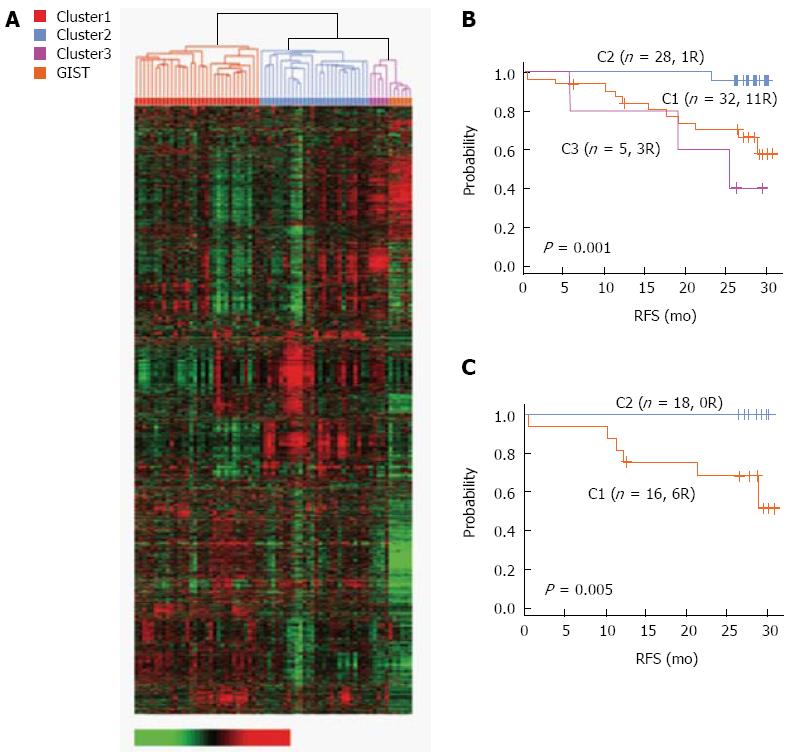
Figure 2 Prognostic expression signatures of genes associated with survival.
A: Hierarchical clustering of gene expression data from 65 gastric cancer and 6 gastrointestinal stromal tumors patients in the Yonsei gastric cancer (YGC) cohort; B: Kaplan-Meier plots of 3 gastric cancer clusters in the YGC cohort; C: Kaplan-Meier plots of stage III patients in 2 clusters (C1 and C2) in the YGC cohort. RFS: Recurrence-free survival.
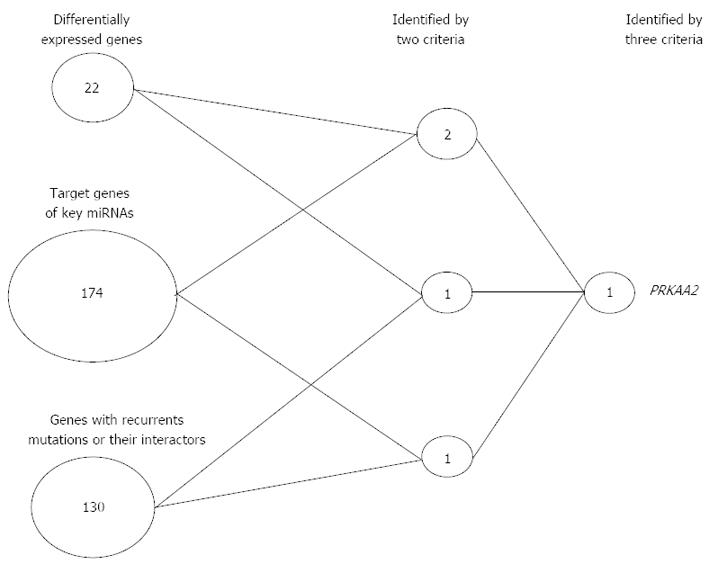
Figure 3 Simple scoring analysis of Asian gastric cancer.
Three criteria were used to select key genes: (1) Genes that were identified in both 5-group and 4-stage differential expression analysis; (2) Target genes of six key differentially expressed miRNAs, where the target genes were predicted by TargetScan (Rs < -0.4, P < 0.05); and (3) Genes with recurrent somatic mutations or their interactors (Ingenuity Pathway Analysis program annotation). Only two genes met two criteria, and PRKAA2 was the only gene that met all three criteria. miRNA: MicroRNA.
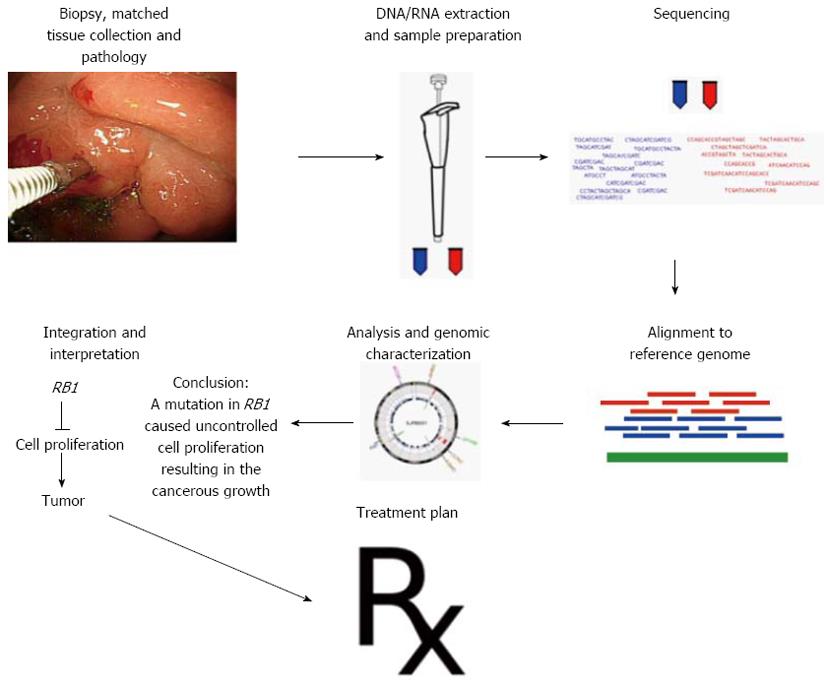
Figure 4 Schematic for genomics-driven cancer medicine where the cancer treatment regimen is adjusted according to each patient’s genome.
- Citation: Lim SM, Lim JY, Cho JY. Targeted therapy in gastric cancer: Personalizing cancer treatment based on patient genome. World J Gastroenterol 2014; 20(8): 2042-2050
- URL: https://www.wjgnet.com/1007-9327/full/v20/i8/2042.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i8.2042